Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity - PubMed
- ️Tue Jan 01 2013
Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity
Christine L Chaffer et al. Cell. 2013.
Abstract
The recent discovery that normal and neoplastic epithelial cells re-enter the stem cell state raised the intriguing possibility that the aggressiveness of carcinomas derives not from their existing content of cancer stem cells (CSCs) but from their proclivity to generate new CSCs from non-CSC populations. Here, we demonstrate that non-CSCs of human basal breast cancers are plastic cell populations that readily switch from a non-CSC to CSC state. The observed cell plasticity is dependent on ZEB1, a key regulator of the epithelial-mesenchymal transition. We find that plastic non-CSCs maintain the ZEB1 promoter in a bivalent chromatin configuration, enabling them to respond readily to microenvironmental signals, such as TGFβ. In response, the ZEB1 promoter converts from a bivalent to active chromatin configuration, ZEB1 transcription increases, and non-CSCs subsequently enter the CSC state. Our findings support a dynamic model in which interconversions between low and high tumorigenic states occur frequently, thereby increasing tumorigenic and malignant potential.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
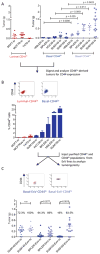
(A) Tumorigenicity of FACS-purified luminal BrCa CD44lo cells or basal BrCa CD44lo and CD44hi cell populations following orthotopic injection into NOD/SCID mice (n ≥ 6/group). (B) Representative FACS plots for CD44 expression and quantification of CD44hi cells generated from luminal or basal CD44lo-derived tumors generated in (A). (C) Basal CD44lo digested tumors from (B) were cultured in vitro to generate ex vivo cell lines (ExV). ExV lines were purified by FACS into CD44lo and CD44hi components and injected orthotopically into NOD/SCID mice (n ≥ 8/group). Tumor incidence displayed as percentages on the graph. Data represented as mean ± SEM. See also Figure S1.
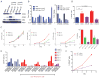
(A) Western blot for markers of the epithelial (CDH1) or mesenchymal (CDH2, VIM) phenotype in immortalized human mammary epithelial cells (HME), HME-flopc and single cell clones derived from HME-flopc population enriched for the CD44lo phenotype (clones F1 and F2) or CD44hi phenotype (clones F3 and F4). (B) qPCR for EMT transcription factors and MIR200B/C in non-transformed CD44lo (HME and HME-flopc-CD44lo) or HME-flopc-CD44hi cells. (C) FACS analysis for the ability of HME-flopc-CD44lo cells to switch to the CD44hi cell state. Cells express a doxycycline (dox) inducible control shRNA (control) or shRNA targeting ZEB1 (sh1 and sh2). -/- no dox, +/- dox on for 8 days then removed for the remaining 8 days, +/+ dox on for the duration of the experiment. Inhibition (%) at day 16 is also shown (*p < 0.0001, **p < 0.0008, compared to (-/-). (D) Purified HME-flopc-CD44lo cells treated with MIR200B/C inhibitors (I) or mimetics (M) to determine effects on switching from CD44lo to CD44hi cell state. Data are mean ± SEM. (E) Purified HME-flopc-CD44lo cells expressing a dox-inducible control shRNA (control) or shRNA targeting ZEB1 (sh1, sh2 or sh3) were analyzed for their ability to switch to the CD44hi state in the presence (+) or absence (-) of dox, and in response to a MIR200C inhibitor (I) or mimetic (M). (F) Transformed HME-flopc-CD44lo cells (with SV40-Early Region and RAS oncoprotein) expressing a dox-inducible shRNA targeting ZEB1 (sh1) were analyzed for conversion to the CD44hi state in the presence (+) or absence (-) of ZEB1-knockdown. Cells were monitored by FACS for 8 days. Data represented as mean ± SEM. See also Figure S2.
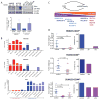
(A) Western blot comparing the expression of ZEB1 in basal BrCa cell lines (HMLER and HCC38) purified for CD44lo or CD44hi subpopulations, and luminal BrCa cell lines (ZR-75-1, T47D, MCF7 and MCF7R). Quantification of differential ZEB1 expression in basal cell lines (n = 4). (B) qPCR assessing ZEB1, MIR200B and MIR200C mRNA expression in BrCa cell lines. (C) Schematic illustrating expression of MIR200 family members and ZEB1 protein expression in basal (CD44lo and CD44hi subpopulations) and luminal BrCa cell lines. (D) Purified CD44lo cells from HMLER or HCC38 and SUM159 basal BrCa cell lines expressing dox-inducible control shRNA (control) or shRNA targeting ZEB1 (sh1 and sh2) were analyzed for tumorigenic potential. Final tumor mass and incidence are represented (n ≥ 5/group). Data represented as mean ± SEM. See also Figure S3
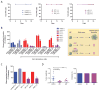
(A) FACS analysis for CD44 expression in purified HME-flopc-CD44hi cells. Cells express a doxycycline (dox) inducible control shRNA (control) or shRNA targeting ZEB1 (sh1 and sh2). -/- no dox, +/- dox on for 8 days then removed for the remaining 8 days, +/+ dox on for the duration of the experiment. The percentage of spontaneously arising CD44lo cells was determined by FACS over a 16 day time period. (B) Purified HME-flopc-CD44hi cells expressing control or shRNA targeting ZEB1 (sh1, sh2 and sh3) were assessed for mammosphere-forming ability with or without dox-induction. Cells were treated with a MIR200C inhibitor (I) or mimetic (M). p<0.001, two-way ANOVA followed by Tukey's multiple comparisons test, *- different to miR-control and miR-200c-M, **- different to miR-control and miR-200c-I (C) Transformed HME-flopc-CD44hi cells expressing control, sh1 or sh2 were assessed for mammosphere formation with or without dox-induction (p<0.0001, one-way ANOVA followed by Tukey's multiple comparisons test, *-different to sh(-)). (D) Transformed HME-flopc-CD44hi cells (control, sh1 and sh2) were purified by FACS and implanted into the fat pad of NOD/SCID mice (n = 8/group). Tumor weight and incidence are shown. Data represented as mean ± SEM. See also Figure S4
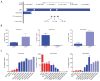
(A) Schematic showing the location of primer sets used for ChIP-qPCR. (B) and (C) ChIP-qPCR for the H3K4me3, H3K27me3 and H3K79me2 histone modifications at the ZEB1 promoter in (B) non-transformed CD44lo or CD44hi cells and (C) luminal CD44lo cells and basal CD44lo and CD44hi sorted populations. Data are mean ± SEM of biological duplicates performed as technical replicates.
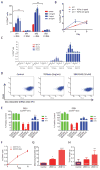
(A) Purified HME-flopc-CD44lo cells expressing dox-inducible shRNA targeting ZEB1 (sh1 and sh2) were monitored by FACS for their ability to switch to the CD44hi state following TGFbeta treatment in vitro. *p<0.0001 - different to control, **p<0.001- different to control (-dox). (B) Transformed HME-flopc-CD44lo cells expressing sh1 targeting ZEB1 were treated with TGFbeta and monitored by FACS for switching to the CD44hi state. (C) Purified CD44lo cells from luminal (MCF7R and ZR-75-1) and basal (HMLER and HCC38) BrCa cell lines monitored by FACS for switching to the CD44hi state following TGFbeta or SB431542 treatment in vitro *p<0.0001 – two-way ANOVA followed by Tukey's multiple comparisons test. (D) Representative FACS plots of HME-flopc-CD44lo cells expressing sh1 or sh2 targeting ZEB1 treated with control (PBS), TGFbeta (2ng/ml) or SB431542 (10μM). (E) ChIP-qPCR for the H3K4me3, H3K27me3 and H3K79me2 histone modifications at the ZEB1 promoter in cells from (D) (*p<0.0001, n = 4, two-way ANOVA followed by Tukey's multiple comparison test, different to control and SB431542). (F-H) MCF7R cells expressing a dox-inducible empty vector (control) or ZEB1 overexpression construct were treated with dox and monitored by FACS for their ability to switch to the CD44hi state (F), for the ability to form tumorspheres in vitro, *p<0.0001, one-way ANOVA and Tukey's multiple comparisons test, different to control (G) and for tumorigenicity in vivo (tumor initiating ability marked as percentages on each bar), *p = 0.03, one-way ANOVA and Tukey's multiple comparisons test, different to Control and ZEB1-lo (H). Data represented as mean ± SEM. See also Figure S6.
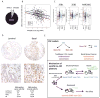
(A) MIR200C accounts for 93% (sd=5%) of mature miRNA in the MIR200BC family, which also includes MIR200B (6%) and MIR429 (1%), so subsequent analysis uses MIR200C to represent the MIR200BC family. (B) Levels of ZEB1 and MIR200C are inversely correlated in basal (p=9.4e-4; r2=0.13), luminal A (p=2.8e-4; r2=0.07) and luminal B (p=6.8e-4; r2=0.12) subtypes (but not Her2). ZEB1 is shown as median-centered values and MIR200C by log2-transformed reads per million mapped reads (RPM). (C) mRNA abundance of ZEB1, ZEB2, and MIR200C by subtype. Asterisks indicate a difference compared to the basal subtype (p<0.05; ANOVA with Dunnett post-hoc): *=p<0.05; **=p<0.01; *=p<0.05. (D) Human BrCa tissue array stained with an antibody targeting ZEB1 (100x and 400x images provided). ** p=0.017, TN compared to Luminal A, Fisher's Exact Test followed by Bonferroni correction for multiple-hypothesis testing. See also Figure S7. (E) Schematic depicting: 1) an alternative CSC model for basal-type BrCa that includes bidirectional conversions between CSCs and non-CSCs, and 2) a model of non-CSC-to-CSC conversion that includes a microenvironmental stimulus acting on non-CSCs harboring bivalent chromatin marks at the ZEB1 promoter enabling a rapid activation of ZEB1 and switch to a CSC state.
Comment in
-
Cancer stem cells: Easily moulded.
Seton-Rogers S. Seton-Rogers S. Nat Rev Cancer. 2013 Aug;13(8):519. doi: 10.1038/nrc3573. Epub 2013 Jul 18. Nat Rev Cancer. 2013. PMID: 23864052 No abstract available.
-
Poised with purpose: cell plasticity enhances tumorigenicity.
Marjanovic ND, Weinberg RA, Chaffer CL. Marjanovic ND, et al. Cell Cycle. 2013 Sep 1;12(17):2713-4. doi: 10.4161/cc.26075. Epub 2013 Aug 19. Cell Cycle. 2013. PMID: 23966153 Free PMC article. No abstract available.
Similar articles
-
Poised with purpose: cell plasticity enhances tumorigenicity.
Marjanovic ND, Weinberg RA, Chaffer CL. Marjanovic ND, et al. Cell Cycle. 2013 Sep 1;12(17):2713-4. doi: 10.4161/cc.26075. Epub 2013 Aug 19. Cell Cycle. 2013. PMID: 23966153 Free PMC article. No abstract available.
-
Lee JY, Park MK, Park JH, Lee HJ, Shin DH, Kang Y, Lee CH, Kong G. Lee JY, et al. Oncogene. 2014 Mar 6;33(10):1325-35. doi: 10.1038/onc.2013.53. Epub 2013 Mar 11. Oncogene. 2014. PMID: 23474752
-
Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, Huang X, Zhao J, Gu B, Zheng GX, Yang AG, Jia LT. Bai WD, et al. Int J Cancer. 2014 Sep 15;135(6):1356-68. doi: 10.1002/ijc.28782. Epub 2014 Mar 3. Int J Cancer. 2014. PMID: 24615544
-
Sato F, Kubota Y, Natsuizaka M, Maehara O, Hatanaka Y, Marukawa K, Terashita K, Suda G, Ohnishi S, Shimizu Y, Komatsu Y, Ohashi S, Kagawa S, Kinugasa H, Whelan KA, Nakagawa H, Sakamoto N. Sato F, et al. Cancer Biol Ther. 2015;16(6):933-40. doi: 10.1080/15384047.2015.1040959. Epub 2015 Apr 21. Cancer Biol Ther. 2015. PMID: 25897987 Free PMC article.
-
MicroRNA control of epithelial-mesenchymal transition in cancer stem cells.
Hao J, Zhang Y, Deng M, Ye R, Zhao S, Wang Y, Li J, Zhao Z. Hao J, et al. Int J Cancer. 2014 Sep 1;135(5):1019-27. doi: 10.1002/ijc.28761. Epub 2014 Feb 19. Int J Cancer. 2014. PMID: 24500893 Review.
Cited by
-
Epigenetic factor competition reshapes the EMT landscape.
Al-Radhawi MA, Tripathi S, Zhang Y, Sontag ED, Levine H. Al-Radhawi MA, et al. Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2210844119. doi: 10.1073/pnas.2210844119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215492 Free PMC article.
-
Cancer stem cells (CSCs), cervical CSCs and targeted therapies.
Huang R, Rofstad EK. Huang R, et al. Oncotarget. 2017 May 23;8(21):35351-35367. doi: 10.18632/oncotarget.10169. Oncotarget. 2017. PMID: 27343550 Free PMC article. Review.
-
Lee G, Auffinger B, Guo D, Hasan T, Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, James CD, Ahmed AU. Lee G, et al. Mol Cancer Ther. 2016 Dec;15(12):3064-3076. doi: 10.1158/1535-7163.MCT-15-0675. Epub 2016 Oct 7. Mol Cancer Ther. 2016. PMID: 27765847 Free PMC article.
-
Xu Y, Baylink DJ, Chen CS, Tan L, Xiao J, Park B, Valladares I, Reeves ME, Cao H. Xu Y, et al. Front Oncol. 2023 Nov 8;13:1286863. doi: 10.3389/fonc.2023.1286863. eCollection 2023. Front Oncol. 2023. PMID: 38023123 Free PMC article.
-
How does multistep tumorigenesis really proceed?
Chaffer CL, Weinberg RA. Chaffer CL, et al. Cancer Discov. 2015 Jan;5(1):22-4. doi: 10.1158/2159-8290.CD-14-0788. Cancer Discov. 2015. PMID: 25583800 Free PMC article. Review.
References
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. - PubMed
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical