An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4 - European Journal of Human Genetics
- ️Jin, Li
- ️Wed Apr 20 2011
Abstract
Y-chromosome Haplogroup O is the dominant lineage of East Asians, comprising more than a quarter of all males on the world; however, its internal phylogeny remains insufficiently investigated. In this study, we determined the phylogenetic position of recently defined markers (L127, KL1, KL2, P164, and PK4) in the background of Haplogroup O. In the revised tree, subgroup O3a-M324 is divided into two main subclades, O3a1-L127 and O3a2-P201, covering about 20 and 35% of Han Chinese people, respectively. The marker P164 is corrected from a downstream site of M7 to upstream of M134 and parallel to M7 and M159. The marker PK4 is also relocated from downstream of M88 to upstream of M95, separating the former O2* into two parts. This revision evidently improved the resolving power of Y-chromosome phylogeny in East Asia.
Similar content being viewed by others
Introduction
The phylogeny of Y-chromosome provides a powerful tool to reconstruct history of human populations and paternal pedigrees.1, 2, 3 The dominant group of Y chromosome in Eastern Eurasia is Haplogroup O-M175, comprising roughly 75% of the Chinese4, 5, 6 and more than half of the Japanese population.7, 8, 9 Albeit the huge population of Haplogroup O, its phylogeny10 is much less adequately resolved than those of Haplogroup R and E, despite the improvements of O tree made during the recent years.10, 11, 12 The most important part of potential interest was the paragroup O3a*-M324(xM121, M159, M164, M7, M134), which comprises a substantial part of Chinese population (typically 15–50% of Han Chinese) but could not be further resolved in the last few years. Fortunately, recent data from HapMap project (http://hapmap.ncbi.nlm.nih.gov/)13 and certain companies show that several single-nucleotide polymorphisms (SNPs) can further distinguish the paragroup O3a*. For example, a novel clade defined by IMS-JST002611 (in short, 002611) may count for up to 21% of the East Asians.4, 8
From the HapMap project, several SNPs were recognized under Haplogroup O-P191 (rs16980601). One of them, rs17269396, was reported as L127.1 on FamilyTreeDNA (https://www.familytreedna.com/advanced-snp-descriptions.aspx) as within O3a-P93. Two further SNPs were reported on DecodeMe (http://demo.decodeme.com/ancestry/male-line-advanced/O), which showed that the derived allele has a substantial proportion in all tested samples belonging to Haplogroup O: rs17276338 (25%) and rs17316007 (P164, 30%), indicating their importance. However, their phylogenetic positions relative to other SNPs were not reported.
The SNP P164 does not agree to the position in the latest Y-chromosome phylogenetic tree as defined in the previous study10 – derivative of SNP M7. Several publicly available individual samples (eg, from 23andMe, https://www.23andme.com/) have derived alleles of both M134 and P164, whereas M134 and M7 are parallel subclades. Another SNP, PK4, defining the clade O2a1a, was first discovered in Pakistan.14 We found that several samples were typed as derivative for PK4 but ancestral for M95 (defining O2a), showing inconsistency with the current tree. Therefore, the position of P164 and PK4 needed to be revised.
Materials and methods
To revise the phylogeny of Haplogroup O, we collected whole blood samples from 361 unrelated Han Chinese male volunteers at Fudan University in Shanghai, with informed consent. The origins of volunteers can be traced back from all over China, although the majority are from East China, that is, Jiangsu, Zhejiang, Shanghai, and Anhui. DNA was extracted from the samples, and then genotyped at M175, M119, P203, M110, M268, P31, M95, M88, PK4, M176, M122, M324, M121, M164, P201, M159, M7, M134, M117, 002611, P164, L127 (rs17269396), KL1 (rs17276338), and KL2 (rs17323322; KL1 and KL2 are named in this study), using the SNaPshot multiplex kit (ABI, Carlsbad, CA, USA) as described before.15 Besides the previously listed primers,10 the primers of newly typed SNPs were designed (Table 1).
Results
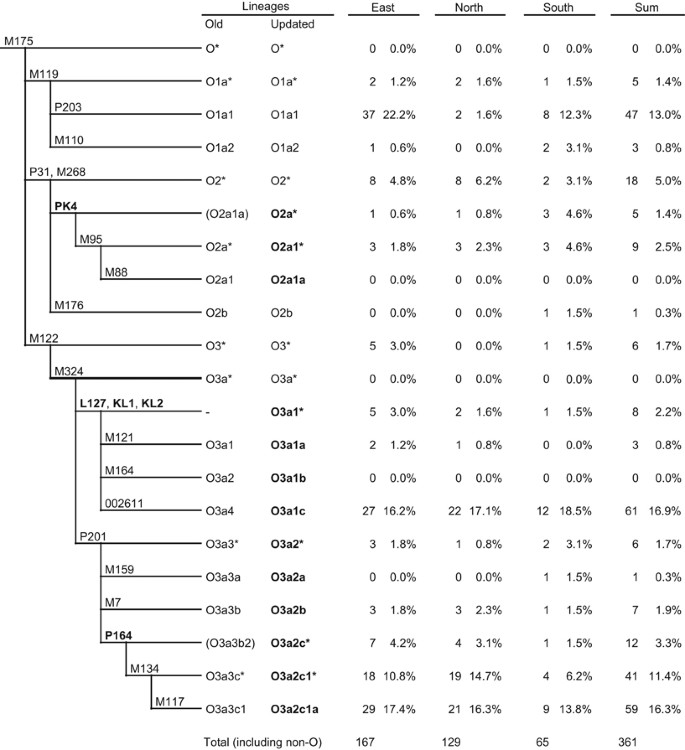
In the updated tree of Haplogroup O (Figure 1), the new lineage names were proposed according to the nomenclature rules.16 The SNPs L127, KL1, and KL2 are found upstream to M121, M164, and 002611, and together define Haplogroup O3a1. The position of P164 is corrected and placed upstream of M134 and downstream of P201; no samples were found sharing P164 and M7 mutations. P201 defines Haplogroup O3a2. The relative position of M300 and M333 to P201, 002611, and L127/KL1/KL2 were not determined in this study, because of lack of positive samples as in the previous study,16 and the relevant lineages can be provisionally named O3a3 and O3a4, respectively. All the samples surveyed that are derivative for M95 have PK4 mutation, whereas there are also (M95−, PK4+) samples indicating that PK4 should be placed upstream of M95, forming an ancient clade in Haplogroup O2.
The updated tree of Haplogroup O and the frequencies of its subgroups in Han Chinese population. The newly located SNPs and renamed lineages are labeled in bold. Samples with derived allele of M164 from our previous studies were typed at L127, KL1, and KL2 in order to locate it in the revised tree. ‘East’ refers to the samples whose origins are from the provinces of Jiangsu, Anhui, Zhejiang, and Shanghai, whereas ‘North’ and ‘South’ refers to the other provinces of which the capitals locate northern or southern to the Line of Qinling Mountains-Huai River, respectively. The samples typed besides Haplogroup O include C-M130 (27, 7.5%), D-M174 (7, 1.9%), G-M201 (1, 0.3%), N-M231 (18, 5.0%), Q-M242 (12, 3.3%), and R-M207 (4, 1.1%).
Discussion
This study shows that L127/KL1/KL2 and P164 are highly informative for separating a substantial part of O3a-M324 samples in China. O3a1-L127 and O3a2-P201 now appear as two main constituent parts of Chinese population (∼20 and ∼35%, respectively), whereas individuals belonging to the newly defined paragroup O3a*-M324(xL127,P201) were not found in this study. This may indicate two main origins of early Han Chinese people, although the exact history can only be discovered through investigation of more populations throughout eastern Eurasia. Although the distribution and age of several clades inside O3a2-P201 (O3a2c1-M134, O3a2c1a-M117, and O3a2b-M7) have been discussed in several studies,5 few were carried out on O3a1-L127. There are no studies about the origin and date of O3a1c-002611 yet, despite its great prevalence and huge population. Concerning the frequency of O3a2*-P201(xM134, M7) in Austronesian-speaking populations (up to 55%),4 it will be interesting to type P164 and M159 on those samples in order to clarify their relationship with the continental populations.
The mutation PK4 separated the former paragroup O2*-M268,P31(xM95,M176) (6.4% in this study) into two parts: PK4− samples are more abundant in Northern and Eastern parts of China, whereas the PK4+ samples are more frequent in the South, showing the same trend with O2a1-M95.5, 17 Previously, the PK4+ samples were only reported in Pakistan, Nepal, and India from two studies,14, 18 in which no (M95+, PK4−) samples were found. Therefore, the relocation of PK4 in this study does not conflict with the result from those two studies. Considering the amount of O2*-M268,P31(xPK4, M176) (5% in this study) and the existence of O2a*-PK4(xM95) samples in China, as well as the incontinuous distribution pattern of O2a1-M95 (frequent in South China, Southeast Asia, and South Asia) and O2b-M176 (abundant in Korea and Japan),7, 9 we may suggest that China was the expansion origin of Haplogroup O2-M268.
With the help of the improved resolution, the newly located SNPs need to be routinely genotyped for the Y-chromosome studies of the East and Southeast Asian populations. The upgraded phylogenetic tree will help to unveil the early history and fine structure of the Asian and Oceanian peoples.
References
Jin L, Su B : Natives or immigrants: modern human origin in East Asia. Nat Rev Genet 2000; 1: 126–133.
Jobling MA, Tyler-Smith C : The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003; 4: 598–612.
Sykes B, Irven C : Surnames and the Y chromosome. Am J Hum Genet 2000; 66: 1417–1419.
Karafet TM, Hallmark B, Cox MP et al: Major east-west division underlies Y-chromosome stratification across Indonesia. Mol BiolEvol 2010; 27: 1833–1844.
Li H, Wen B, Chen SJ et al: Paternal genetic affinity between western Austronesians and Daic populations. BMC Evol Biol 2008; 8: 146.
Su B, Xiao JH, Underhill P et al: Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 1999; 65: 1718–1724.
Hammer MF, Karafet TM, Park H et al: Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet 2006; 51: 47–58.
Nonaka I, Minaguchi K, Takezaki N : Y-chromosomal binary haplogroups in the Japanese population and their relationship to 16 Y-STR polymorphisms. Ann Hum Genet 2007; 71: 480–495.
Xue YL, Zejal T, Bao WD et al: Male demography in East Asia: a north-south contrast in human population expansion times. Genetics 2006; 172: 2431–2439.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Deng W, Shi BC, He XL et al: Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J Hum Genet 2004; 49: 339–348.
Shi H, Dong YL, Wen B et al: Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet 2005; 77: 408–419.
The International HapMap Consortium: A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Firasat S, Khaliq S, Mohyuddin A et al: Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan. Eur J Hum Genet 2007; 15: 121–126.
Inagaki S, Yamamoto Y, Doi Y et al: Typing of Y chromosome single nucleotide polymorphisms in a Japanese population by a multiplexed single nucleotide primer extension reaction. Leg Med (Tokyo) 2002; 4: 202–206.
The Y-Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Su B, Deka R, Xiao C et al: The origin and genetic affinity of Sino-Tibetan speaking populations. Am J Human Genetics 1999; 65: 2260.
Fornarino S, Pala M, Battaglia V et al: Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation. BMC Evol Biol 2009; 9: 154.
Acknowledgements
This research was supported by grants from the National Outstanding Youth Science Foundation of China (30625016) and National Science Foundation of China (30890034). Genographic Consortium members are the following: Theodore G. Schurr (University of Pennsylvania, Philadelphia, PA, USA), Fabricio R.Santos (Universidade Federal de Minas Gerais, Belo Horizonte, Brazil), Lluis Quintana-Murci (Institut Pasteur, Paris, France), Jaume Bertranpetit, David Comas (Universitat Pompeu Fabra, Barcelona, Spain), Chris Tyler-Smith (The Wellcome Trust Sanger Institute, Hinxton, UK), Pierre A. Zalloua (Lebanese American University, Chouran, Beirut, Lebanon), Elena Balanovska, Oleg Balanovsky (Russian Academy of Medical Sciences, Moscow, Russia), R. John Mitchell (La Trobe University, Melbourne, Australia), Li Jin (Fudan University, Shanghai, China), Himla Soodyall (National Health Laboratory Service, Johannesburg, South Africa), Ramasamy Pitchappan (Madurai Kamaraj University, Madurai, India), Alan Cooper (University of Adelaide, Adelaide, Australia), Lisa Matisoo-Smith (University of Auckland, Auckland, New Zealand), Ajay K. Royyuru, Daniel E. Platt, Laxmi Parida (IBM, Yorktown Heights, New York, NY, USA), Jason Blue-Smith, David F. Soria Hernanz, and R. Spencer Wells (National Geographic Society, Washington, D.C., USA).
Author information
Authors and Affiliations
State Key Laboratory of Genetic Engineering and Ministry of Education, Key Laboratory of Contemporary Anthropology, School of Life Sciences and Institutes of Biomedical Sciences, Fudan University, Shanghai, China
Shi Yan, Chuan-Chao Wang, Hui Li, Shi-Lin Li & Li Jin
Group of Computational Genetics, CAS-MPG Partner Institute for Computational Biology, SIBS, CAS, Shanghai, China
Shi Yan & Li Jin
Authors
- Shi Yan
You can also search for this author in PubMed Google Scholar
- Chuan-Chao Wang
You can also search for this author in PubMed Google Scholar
- Hui Li
You can also search for this author in PubMed Google Scholar
- Shi-Lin Li
You can also search for this author in PubMed Google Scholar
- Li Jin
You can also search for this author in PubMed Google Scholar
Consortia
The Genographic Consortium
- Theodore G Schurr
- , Fabricio R Santos
- , Lluis Quintana-Murci
- , Jaume Bertranpetit
- , David Comas
- , Chris Tyler-Smith
- , Pierre A Zalloua
- , Elena Balanovska
- , Oleg Balanovsky
- , R John Mitchell
- , Li Jin
- , Himla Soodyall
- , Ramasamy Pitchappan
- , Alan Cooper
- , Lisa Matisoo-Smith
- , Ajay K Royyuru
- , Daniel E Platt
- , Laxmi Parida
- , Jason Blue-Smith
- , David F Soria Hernanz
- & R Spencer Wells
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Yan, S., Wang, CC., Li, H. et al. An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4. Eur J Hum Genet 19, 1013–1015 (2011). https://doi.org/10.1038/ejhg.2011.64
Received: 19 October 2010
Revised: 23 February 2011
Accepted: 09 March 2011
Published: 20 April 2011
Issue Date: September 2011
DOI: https://doi.org/10.1038/ejhg.2011.64