Dysregulation of the C/EBPα Differentiation Pathway in Human Cancer
- ️Mon Feb 07 2005
Abstract
While much is known about aberrant pathways affecting cell growth and apoptosis, our understanding of another critical step of neoplastic transformation, differentiation arrest, remains poor. The differentiation-inducing transcription factor CCAAT enhancer binding protein alpha (C/EBPα) is required for proper control of adipogenesis, glucose metabolism, granulocytic differentiation, and lung development. Studies investigating the function of this protein in hematopoietic malignancies as well as in lung and skin cancer have revealed numerous ways how tumor cells abrogate C/EBPα function. Genetic and global expression analysis of acute myeloid leukemia (AML) cases identifies C/EBPα-deficient AML as a separate entity yielding novel classification schemes. In patients with a dysfunctional C/EBPα pathway, targeted therapies may overcome the block in differentiation, and in combination with conventional chemotherapy, may lead to complete eradication of the malignant clone. Overall, a better understanding of the mechanisms of how C/EBPα dysregulation participates in the neoplastic process has opened new gateways for differentiation biology research.
INTRODUCTION
Disruption of normal differentiation is a critical component of tumorigenesis, but the genetic mechanisms of this process are still poorly understood. Cells must be capable to enter a quiescent state in a precisely regulated manner, such as during terminal differentiation to maintain tissue homeostasis, or in response to DNA damage or oncogenic stress. Lineage-specific transcription factors control both of these processes and thereby play a pivotal role in the determination of cellular fate.1 C/EBPα is a critical transcription factor that controls tissue-specific gene expression and proliferation arrest, thereby coordinately regulating two essential features of terminally differentiated cells. Recent studies demonstrate that C/EBPα indeed acts as a bona fide tumor suppressor in a number of tumor types,2,3 supporting the view that disruption of normal differentiation and its uncoupling from cell cycle arrest are key components in the development of cancer.4 The present review will summarize our current understanding of the role of C/EBPα in neoplasia.
PHYSIOLOGICAL FUNCTION OF C/EBPα
The C/EBP Family of Transcription Factors
C/EBP proteins form a family of basic region leucine zipper (bZIP) transcription factors that includes six members with related sequences and functions (α, β, γ, δ, ε and ζ).5–10 C/EBPs are modular proteins consisting of two transactivation domains, a dimerization leucine zipper region, and a DNA-binding domain. Dimerization results in approximation of the basic amino acid residues and is a prerequisite for DNA binding. When the phasing between the basic domain and leucine zipper is altered by nucleotide insertions or deletions as described below in cases of C/EBPα mutations in AML, DNA-binding is altered or eliminated. The x-ray structure of the C/EBPα protein-DNA interaction was recently solved, and several critical basic region residues were identified.11
Structure and Function of C/EBPα
The C/EBPα (C/EBPα) gene is localized to chromosome 19q13.1, is intronless, and two protein isoforms are generated from two inframe AUG codons on the mRNA (Fig 1C). The shorter form, known as p30, contains the same carboxyl terminus as the full-length form, known as p42, but lacks the amino-terminal 117 amino acids.12 The transactivation domains TE-I and TE-II13 (regions 1 to 3 and 5 to 614) that interact with the transcriptional apparatus are only present in the p42 isoform, while the domains required for interaction with other transcription factors and the chromatin remodeling complex are present in both isoforms.13,14
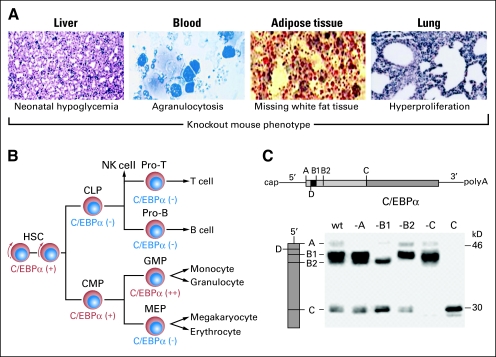
Fig 1.
Physiological function of CCAAT enhancer binding protein alpha (C/EBPα). (A) Major sites of tissue expression of C/EBPα include the liver, the hematopoietic system, white adipose tissue, and the lung. Accordingly, mice with targeted disruption of C/EBPα (C/EBPα-knockout mice) display perinatal hypoglycemia, absence of granulocytes, lack of white adipose tissue, and hyperproliferation of interstitial lung tissue. Modified from Zhang et al24, Yang et al24a and Flodby et al23 (with permission). (B) Within the hematopoietic system, C/EBPα is expressed at lower levels in hematopoietic stem cells (HSCs), and its expression increases progressively during granulocytic differentiation via common myeloid progenitor cells (CMP) and granulocyte-macrophage progenitor cells (GMP). C/EBPα is not detectable in megakaryocyte-erythrocyte progenitors (MEP), common lymphoid progenitors (CLP), or more mature lymphoid cells. Modified from Akashi et al with permission.113 (C) The C/EBPα mRNA is comprised of a cap sequence and 5′ untranslated region (UTR), the C/EBPα coding sequence derived from a single exon, and the 3′ UTR, which is followed by the poly A sequence (upper panel). The letters A, B1, B2, C, and D designate five ATG sites of translational start sites which are highly conserved across the six different species depicted with the exception of the A site, which is missing in the human species (middle panel). Translation from all sites except D produce a specific C/EBPα isoform that can be detected by Western blotting, and omission of each site (ie, −A) leads to the loss of the specific isoform. The major C/EBPα isoforms are produced from the B1 and C sites and are 42 kD and 30 kD in size. From Calkhoven et al with permission.114 NK, natural killer; Pro-T, T-cell progenitors; Pro-B, B-cell progenitors; wt, wild type.
C/EBPα is expressed at high levels in a specific subset of tissues and cell types, such as liver, adipose tissue, myeloid cells, and airway epithelial cells.15,16 In these cells, C/EBPα directly activates transcription from lineage-specific gene promoters (ie, serum albumin, AP2 and stearoyl-coenzyme A desaturase (SCD1), granulocyte colony-stimulating factor receptor, or surfactant proteins and CC10). Direct contact with the basal transcriptional apparatus (TBP/TFIIB), interaction with histone acetyltransferases (CBP/p300), and recruitment of chromatin-remodelling complexes (SWI/SNF) have all been implicated in the activation of lineage-specific genes by C/EBPα. The puzzling ability of C/EBPα to direct cellular differentiation in a tissue-specific manner is thought to depend on the presence of specific collaborating transcription factors. In adipocytes C/EBPα mediates growth arrest and terminal differentiation in concert with peroxisome proliferator-activated receptor gamma (PPARγ).17 In the hematopoietic system, the interplay between C/EBPα and GATA factors is a similar example18,19 while in the lung, the tissue-specific regulatory network includes thyroid transcription factor-1, FoxA2 (also called HNF3β), and C/EBPα.20
Much has been learned about the role of C/EBPα in tissue development and differentiation from the study of C/EBPα−/− knockout mice21–23 (Fig 1A). These mice die at birth as a result of severe hypoglycemia, and they show disturbed hepatic tissue architecture with proliferative acinar formations resembling pseudoglandular hepatocellular carcinoma. In addition, C/EBPα knockout mice display lung abnormalities such as a striking hyperproliferation of type II pneumocytes. Moreover, they show a complete lack of granulocytes, while blasts accumulate in the bone marrow, suggestive of an early block in myeloid maturation.24
C/EBPα As a Master Regulator of Hematopoietic Differentiation
The maturation of hematopoietic precursors from hematopoietic stem cells (HSC) to differentiated cells is associated with two fundamental processes: reduction in self-renewal potential and stepwise acquisition of a specific lineage identity.1,25 Transcription factors are master regulators of these graduated processes, and aberrations in their regulation/function can lead to loss of lineage identity, increased infidelity, lineage reprogramming, and malignant transformation. The formation of myeloid cells is orchestrated by a small number of critical transcription factors, such as PU-1, C/EBPα/β and δ, IRF-8, RUNX1, SCL, and GFI1. This is well reflected by severe defects seen in myeloid development of mice with targeted deletions of genes encoding these key transcription factors. C/EBPα regulates the expression of many myeloid genes, including genes encoding growth factor receptors (granulocyte colony-stimulating factor, macrophage colony-stimulating factor, granulocyte-macrophage colony-stimulating factor) and the so-called secondary granule proteins (ie, lactoferrin).26–28 C/EBPα has also recently been implicated in the regulation of the microRNA miR223, with C/EBPα-mediated upregulation of miR223 leading to granulocytic differentiation in a myeloid differentiation model.29 Also, the transcription of genes that are normally expressed in other lineages is actively suppressed, driving cells away from granulocytic differentiation toward other differentiation pathways. C/EBPα in the hematopoietic system is expressed in HSCs, myeloid progenitors and granulocytes, but not macrophages.30 C/EBPα-deficient mice have normal numbers of common myeloid progenitors (CMPs) but lack granulocyte-monocyte progenitors (GMPs) and all subsequent granulocytic stages (Fig 1B). Conditional deletion of C/EBPα in GMPs allows for normal granulopoiesis in vitro, indicating that C/EBPα is not required for granulopoietic differentiation beyond the GMP stage.30 C/EBPα also controls stem cell self-renewal properties as C/EBPα-deficient HSCs show increased competitive repopulation activity in mouse transplant studies.30 The increased self-renewal potential of C/EBPα−/− HSCs may, at least in part, be due to elevated expression of the polycomb gene Bmi-1. The mechanism of how altered C/EBPα function ultimately leads to neoplasia may be through failure to induce differentiation in progenitor cells leading to a prolonged and aberrant proliferation phase in these cells followed by random accumulation of additional genetic events and full transformation.
C/EBPα and Proliferation Arrest
C/EBPα possesses the ability to arrest cell proliferation in addition to its ability to drive terminal differentiation and thereby functions as a master switch between proliferating uncommitted and cell cycle–arrested differentiated cells.3,31,32 The mechanisms of C/EBPα-mediated growth arrest include interaction with CDK2/CDK4, upregulation of p21waf1/cip1 and the SWI/SNF chromatin-remodelling complex33–35 as well as direct inhibition of the E2F complex.36,37 Downregulation of c-myc is one of the critical steps of C/EBPα-mediated E2F repression and is required for granulocytic differentiation.31 Point mutations of the basic region of C/EBPα abrogating C/EBPα-E2F complex formation (BRM-5: Y285A and BRM-2 I294A/R297A) interfere with differentiation in both myeloid and adipocytic differentiation.37 These mutants lack growth inhibitory properties, possibly through diminished binding of E2F and/or abolished DNA-binding, as suggested by structural analyses. Interestingly, the shorter p30 isoform lacks E2F-repression and thus lacks antimitotic activity in adipogenic and granulocytic cell types.
ROLE OF C/EBPα IN HEMATOPOETIC MALIGNANCIES
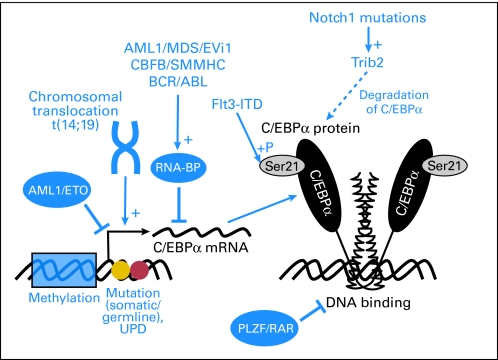
Deregulation of C/EBPα plays a role in a number of hematologic malignancies, where C/EBPα has been demonstrated to be mutated, involved in chromosomal translocations, or suppressed both at the transcriptional and posttranscriptional level, resulting in different mechanisms of oncogenic events (Fig 2).
Fig 2.
Pathogenetic mechanisms of CCAAT enhancer binding protein alpha (C/EBPα) dysfunction. C/EBPα function is disturbed at various levels in acute leukemias. Promoter methylation leads to silencing of promoter activity and decreased expression of C/EBPα. The fusion protein AML1-ETO suppresses C/EBPα transcription via disturbed autoregulation. Chromosomal translocations involving the immunoglobin H locus and the CEBPA gene (t14;19) occur in B-cell precursor acute lymphoblastic leukemia and have been described to induce aberrant C/EBPα expression in lymphoid cells. Other translocations which are associated with AML (AML1/MDS/Evi1 and CBFB/SMMHC) or chronic myelogenous leukemia (CML; BCR-ABL) have been demonstrated to increase the expression of specific RNA binding proteins which can block proper C/EBPα translation from an intact mRNA. In addition, posttranslational modifications such as Serine-21 phosphorylation by oncogenic Flt3-ITD as well as impairment of C/EBPα DNA binding by PLZF-RAR results in dysfunction of C/EBPα and a block of granulocytic differentiation. Finally, C/EBPα protein degradation is induced by Trib2 whose expression is increased by Notch1 mutations in AML. CBFB, core-binding factor β; BP, RNA-binding protein; UPD, uniparental disomy.
Somatic Gene Mutations
Since C/EBPα is essential for granulocytic differentiation and cells from patients with acute myeloid leukemia harbor a differentiation block resulting in an excess of myeloid blasts, it was hypothesized that C/EBPα may be absent or mutated in myeloid leukemias. A screen for mutations within the C/EBPα gene coding sequence revealed that C/EBPα was mutated in approximately 7% of all patients with AML,2 and the results have since been reproduced and extended by other groups38–47 (Fig 3A and B). Intriguingly, mutations were predominantly found within the M1 and M2 FAB morphological subtypes, and no mutations were found in patients with a t(8;21), inv16, or t(15;17) karyotypic abnormality.2 When analyses were performed in patients with a normal karyotype (cytogenetically normal [CN]), 10% to 15% of cases harbor a C/EBPα mutation.2,42 Clinically, C/EBPα mutations are associated with lower leukocyte counts and lactate dehydrogenase levels and with aberrant expression of T-cell surface markers such as CD7 at presentation.48 Moreover, C/EBPα mutations are confined to AML and high-risk myelodysplastic syndrome and have not been found in other diseases. Genome-wide gene expression analysis of blasts from AML patients using unsupervised clustering revealed that C/EBPα mutant cases cluster together, suggesting that they share similar gene expression signatures.49C/EBPα mutations (15% of all CN-AML cases) do not segregate with any other known mutations, such as NPM1, Flt3 or MLL mutations.50
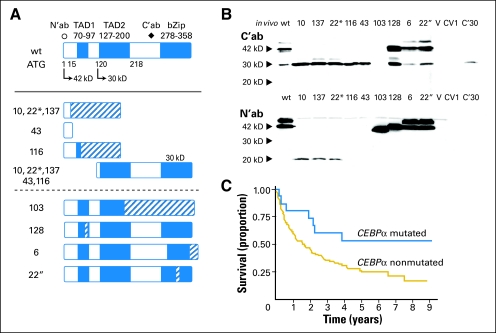
Fig 3.
CCAAT enhancer binding protein alpha (C/EBPα) mutations and their effect on prognosis in acute myeloid leukemia (AML). (A) and (B) Mutations of the coding sequence of C/EBPα have been described in approximately 15% of patients with AML and a normal karyotype by conventional cytogenetics. Two thirds of these patients harbor N-terminal frameshift mutations and C-terminal in-frame insertion/deletion mutations, leading to a nonfunctional 20 kD isoform and increased 30 kD isoform expressed from the N-terminally mutated allele as well as a dysfunctional 42 kD isoform expressed from the C-terminally mutated allele (ie, patient 2”). Please see Pabst et al2 for more details. (C) Retrospective survival analysis of patients with AML harboring C/EBPα mutations or no C/EBPα mutations showing superior overall survival of patients with C/EBPα mutations. Data from Preudhomme et al38. N′ab, antibody recognizing N-terminal portion; TAD, transactivation domain; C'ab, antibody recongnizing the C-terminal domain; bZip, basic zipper region; wt, wild type.
Retrospective studies show that patients with C/EBPα mutant AML have an improved prognosis38,41,42,45 (Fig 3C). Whether this is counteracted by the negative impact of concurrent Flt3-ITD mutations is controversial.38,42 For Flt3-TKD mutations, additional improvement of event-free survival has been described for patients with C/EBPα mutations.51 Prospective validation of the prognostic impact of C/EBPα mutational status is awaited to determine the clinical impact of C/EBPα mutation analysis. The C/EBPα mutations found in AML are typically biallelic and involve frameshift mutations of the 5′ region of the coding sequence on one allele and in-frame insertion/deletion mutations within a narrow 3′ region on the other allele, leading to so-called N-terminal and C-terminal mutations of the protein, respectively52 (Fig 3A and B). Sole N-terminal, sole C-terminal, and other mutations were found in 11.4%, 9%, 12.6% of cases.52 C-terminal mutations are usually located at the junction between the basic region and the leucine zipper coding sequence resulting in impaired alignment with the major groove of DNA and disturbed DNA binding by the mutant protein as well as altered dimerization with its partner proteins.53 N-terminal mutations lead to a nonfunctional 20 kD C/EBPα isoform or a shorter additional isoform while the physiological 30 kD isoform generated from an internal translational start site is always preserved2 (Fig 3A and B). The ratio of the 30 kD isoform and the residual 42 kD full-length isoform generated from the nonmutated allele is increased in the mutant cells, and functional studies have revealed that the 30 kD isoform shows significantly reduced binding to and transactivation of C/EBPα target promoters.2
The exact mechanism of how the C/EBPα mutants inhibit granulocytic differentiation is not entirely clear at present. N-terminal mutants have been described to exert dominant-negative effects on the unmutated 42 kD C/EBPα protein isoform. This may be due a change of the ratio of p30 and p42 homo- and heterodimers. Alternatively, an imbalance between p42 and p30 may affect heterodimerization with other C/EBP members and thus interfere with C/EBP-induced cell cycle arrest and differentiation. The mechanism of C/EBPα-mutant leukemogenesis has been addressed in homozygous knock-in mice that express p30 C/EBPα but lack p42 C/EBPα completely. These mice exhibit an initial phase of neutropenia and go on to develop overt AML with full penetrance.54 Blasts display increased proliferative activity and harbor a normal karyotype similar to C/EBPα-mutant human AML. Granulocyte-macrophage progenitor cells (GMPs) are still formed in preleukemic mice showing that p30 is sufficient to induce commitment of common myeloid progenitors (CMPs) toward GMPs.54 This is in contrast to C/EBPα knockout mice, which lack all C/EBPα isoforms completely and fail to support the transition from CMPs to GMPs. While bone marrow–derived myeloid progenitors from heterozygous wild-type/p30 mice showed only mildly increased proliferative activity compared with wild-type progenitors, p30/p30 homozygous GMPs showed high proliferative activity, high rates of frequent spontaneous immortalization, and increased self-renewal. Therefore, p30 C/EBPα is required and sufficient for commitment of CMPs to the myeloid lineage, whereas p42 C/EBPα is required for cell cycle arrest and the tumor-suppressing functions in myeloid progenitors. Together with the fact that C/EBPα-mutant AML patients also retain expression of p30 C/EBPα, these data suggest that residual expression is required for commitment of HSC toward the myeloid lineage and for the development of AML55 similar to how AML develops in PU.1-hypomorphic but not PU.1 knockout mice.56 This concept is supported by the observation that C/EBPα knockout cells are unable to give rise to a myeloid disease on transduction with BCR-ABL but instead develop an acute erythroblastic leukemia, while wild-type C/EBPα cells give rise to a chronic myelogenous leukemia (CML)–like myeloid disease.55
Germline C/EBPα Mutations
Germline mutations of C/EBPα have been described and are typically N-terminal mutations that are then joined by a somatic C-terminal mutation on the other allele.44,57 The first report described a germline 212delC N-terminal C/EBPα mutation in two siblings and their father with AML.44 Additionally, a 36bp C-terminal C/EBPα gene duplication was found in the leukemic samples of one of the siblings.44 The next report described five individuals with familial AML harboring a germline 217insC mutation.57 Two of these had additional C-terminal insertions at nucleotide 1071. A third report identified a Japanese family with an N-terminal germline mutation and an additional C-terminal mutation in two individuals developing AML.58 The germline mutations were N-terminal mutations in all cases succeeded by a C-terminal somatic mutation in the leukemic blasts suggesting that the combination of these mutations induces AML but is still tolerated by myeloid cells, while a biallelic combination of two C-terminal or two N-terminal mutations is deleterious to the cells and would induce apoptosis instead of AML.
Involvement in Chromosomal Translocations (B-precursor acute lymphoblastic leukemia)
In addition to harboring C/EBPα mutations, the C/EBPα gene as well as other C/EBP members have been demonstrated to be involved in t(14;19)(q32;q13) chromosomal translocations in B-cell precursor acute lymphoblastic leukemia (BP-ALL).59,60 The exact consequences of these translocations are unclear. However, high C/EBPα mRNA and protein were demonstrated in several of these BP-ALL cells, which normally do not express any C/EBPα.59 These observations suggest opposing functions of C/EBPα dysregulation in myeloid and lymphoid leukemogenesis, with C/EBPα serving as an oncogene in BP-ALL.59 This is reminiscent of PU.1 overexpression found in murine erythroleukemias (Friend′s erythroleukemia).61
Uniparental Disomy Involving the C/EBPα Gene
Uniparental disomy (UPD) refers to large regions of acquired homozygosity resulting from mitotic recombination allowing malignant cells to multiply gene mutations and to dispose of wild-type alleles obtaining a selective growth advantage over cells without UPD. UPD was found by genome-wide single nucleotide polymorphism analysis to occur in approximately 20% of AML cases.62 Moreover, the authors detected homozygous mutations at four distinct gene loci that have been associated with AML (WT1, FLT3, C/EBPα, and RUNX1). In line with these data, an AML case of UPD on chromosome 19 with a homozygous C-terminal C/EBPα mutation has been reported.63 These results imply that the gene mutation precedes mitotic recombination and depict another means of AML blasts to dispense of the C/EBPα wild-type allele as has already been described for the coincidence of N- and C-terminal C/EBPα mutations.
Suppression of C/EBPα Transcription
Silencing of tumor suppressor genes by hypermethylation is common in human cancers including leukemias.64,65 Methylation of the C/EBPα core promoter (−600 to ATG) was found to be an infrequent event in AML,66 but more recent studies find that methylation of distal promoter elements (−1600 to-600 from ATG) occurs in 30% of AML patients.67 C/EBPα promoter methylation was present in six of 10 cases with undetectable C/EBPα mRNA expression and was associated with activating Notch1 mutations and aberrant T-cell marker expression.48 Activation of Notch1 led to overexpression of Trib2, a protein which was found to decrease p42 C/EBPα protein—possibly through enhanced proteasomal degradation—and to induce AML in mice.68 Interestingly, these AML cases also clustered together with the C/EBPα mutant samples in the previously mentioned microarray analysis.49 C/EBPα mutations have not been described in cases of t(8;21) or inv16 AML. While inversion 16 cases show hypermethylation of the C/EBPα promoter,69 t(8;21) cases do not. In these cases, C/EBPα mRNA is downregulated by the oncogene AML1-ETO by a mechanism that is still incompletely understood but appears to involve an altered self-stimulatory transcriptional loop.2
Suppression of Protein Translation
Two main mechanisms of inhibition of C/EBPα protein translation have been reported. C/EBPα translation is inhibited in blasts from patients with CML myeloid blast crisis70 via an RNA binding protein, hnRNP-E2, which binds to the upstream open reading frame of the C/EBPα gene and inhibits translation of C/EBPα mRNA. hnRNP-E2 is induced by the oncogenic kinase BCR-ABL via a MAPK-dependent mechanism.71 Downregulation of hnRNP-E2 by the BCR-ABL inhibitor imatinib restored both C/EBPα protein expression and granulocytic differentiation of the blasts.70 Calreticulin, another RNA binding protein, has been demonstrated to be upregulated and to inhibit C/EBPα translation in AML subtypes with CBFB-SMMHC and AML1-MDS-EVI1 translocations.72,73
Posttranslational Modification
Inhibition of C/EBPα activity can also occur through posttranslational modifications. C/EBPα is phosphorylated on serine 21 shifting the ability of C/EBPα to induce differentiation from a granulocytic toward a monocytic phenotype. The oncogenic kinase Flt3-ITD (internal tandem duplication) phosphorylates C/EBPα on ser21 thereby inhibiting granulocytic differentiation.74 Another oncogenic protein, PLZF-RARA, has been shown to interfere with C/EBPα transactivating activity on the granulocyte colony-stimulating factor receptor promoter.75 In addition, Ubc9 has been described to modify C/EBPα transactivation activity.76,77
THE ROLE OF C/EBPα IN AIRWAY EPITHELIAL DIFFERENTIATION AND LUNG CANCER
C/EBPα As a Master Regulator of Airway Epithelial Differentiation
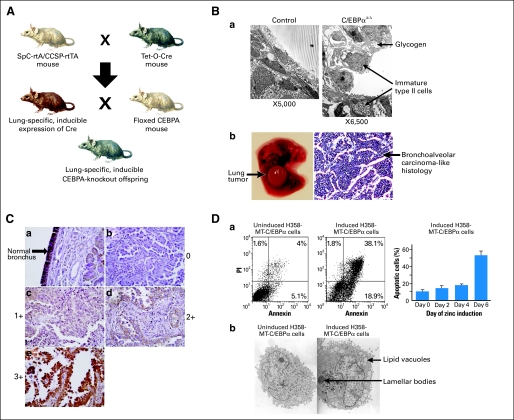
A number of transcription factors such as thyroid transcription factor-1, FoxA2, and C/EBPα control the intricate process of lung development and airway epithelial differentiation78,20,79,80 Induction of C/EBPα expression occurs in close temporal proximity to the initiation of cellular differentiation in the developing lung. In the adult lung, C/EBPα is expressed in both bronchial epithelial Clara cells as well as type II alveolar cells. C/EBPα can regulate the expression of a number of lung-specific genes, such as Clara cell secretory protein (CCSP), surfactant protein A (SP-A), and surfactant protein D (SP-D). CCSP, SP-A and SP-D.81 A conditional mouse model of C/EBPα deletion directed to the respiratory epithelium was developed using tissue-specific tetracycline-regulated cre-recombinase mediated excision of C/EBPα (Fig 4).82 In these mice, in utero loss of C/EBPα leads to respiratory arrest and death soon after birth due to a differentiation arrest of type II alveolar cells associated with proliferation of immature type II alveolar cells, causing epithelial cell expansion and loss of airspace. Expression analysis of knockout versus wild-type lungs demonstrated that the expression of genes characteristic of differentiated pulmonary cells (AQ5, SPA, ABCA3) were downregulated, and genes associated with proliferation (Rgs2, G0s2), tumor progression (Tiam1, Pcsk6) and cell survival (TGFbeta2, Gli1) were upregulated in CEBPα-deficient cells. Similar findings were noted in a study of Martis et al.83 These observations suggest that modulating C/EBPα function in the lung may be useful for the treatment of different forms of respiratory distress syndrome. Interestingly, C/EBPα is detected at the level of bronchoalveolar stem cells (unpublished data), a cell population that has been recently linked to adenocarcinoma initiation. Postnatal excision of C/EBPα is feasible in these inducible mice, allowing the study of the effects of C/EBPα downregulation in predisposing to lung tumorigenesis. Our data obtained on two different lung conditional models (adult SPC and CC10-driven excision) demonstrate significantly increased numbers of lung adenomas and adenocarcinomas that develop on excision of C/EBPα in the adult lung epithelium, supporting the hypothesis that C/EBPα is a lung tumor suppressor (unpublished data). Our data also indicate that Gli-1, a transcriptional effector of the sonic hedgehog pathway, is highly active only in C/EBPα-deleted pulmonary cells that might represent the cells of origin of the lung adenocarcinomas found in our murine models.
Fig 4.
CCAAT enhancer binding protein alpha (C/EBPα) in lung cancer. (A) Generation of tissue-specific, inducible C/EBP-knockout mice. (Ba) Electron microscopy of airway epithelium in a lung-specific C/EBPα-knockout mouse demonstrates hyperproliferation of immature alveolar cells as well as intracellular accumulation of glycogen. (Bb) Histology of lung tumors in C/EBPα-knockout mice demonstrates bronchoalveolar carcinoma-like histology. (C) Immunohistochemical studies demonstrate downregulation of C/EBPα in non–small-cell lung cancer. The panels shown demonstrate strong expression of C/EBPα in normal bronchial epithelium (a) and show non–small-cell lung tumors with different degrees of expression (panels b, 0; c, 1+; d, 2+; e, 3+). (D) In vitro studies demonstrate growth arrest, (a) apoptosis, and (b) and cellular differentiation in lung cancer cell lines as a result of C/EBPα expression in H358 lung cancer cells. Reprinted with permission from Halmos et al,84 Costa et al,85 and Basseres et al.82 H358-MT, H358 cells transfected with a zinc-inducible metallothionein promoter-driven expression plasmid.
C/EBP α As a Lung Cancer Tumor Suppressor
These observations prompted us to examine the role of C/EBPα in human lung cancer.84 A screen of established lung cancer cell lines demonstrated that C/EBPα expression was undetectable or low in 24 of 30 cell lines examined, and immunohistochemical studies confirmed that C/EBPα expression is lost in more than half of all primary tumor specimens. C/EBPα expression is lost more commonly in squamous cell cancers, and there is a trend toward a higher frequency of loss in poorly differentiated and more advanced cancers, suggesting the biologic role of C/EBPα in differentiation and tumor progression, but no prognostic role for C/EBPα expression was found.85 In conditional cell line models, induction of C/EBPα led to significant growth reduction attributable to proliferation arrest, morphological changes suggestive of airway epithelial differentiation, and ultimately apoptosis. A transcriptional profiling study performed using these models identified the forkhead family member and differentiation gene, FoxA2, as directly regulated and induced by C/EBPα in lung cancer cells. FoxA2 appears to be a major mediator of the growth-inhibiting action of C/EBPα.86 This finding is particularly intriguing since FoxA2 is a critical transcription factor in foregut development and in airway epithelial differentiation and FoxA2 itself is a candidate tumor suppressor in lung cancer.
The chromosomal region including C/EBPα was found deleted in 50% of non–small-cell lung cancers suggestive of the presence of a tumor suppressor at this locus.87 We completed sequencing of 31 established lung cancer cell lines and 50 primary lung cancers and identified mutations of C/EBPα in only three of 81 specimens examined.88 Sequencing of the C/EBPα gene by another group in 36 non–small-cell lung cancers failed to identify any coding sequence mutations.39 These findings suggest that mutations of C/EBPα are rare events in lung cancers and that C/EBPα deactivation occurs through alternative mechanisms. The promoter and exonic regions of C/EBPα meet the definition of a CpG island spanning almost 3.4 kb. Interestingly and similar to studies in hematopoietic cells, no methylation was found in the core promoter region. Conversely, hypermethylation of an upstream promoter region of the C/EBPα gene (−1422 to −896 relative to the transcriptional start site) is a very common event in non–small-cell lung cancers (detected in approximately 65% of primary lung tumors) and appears to be the major mechanism for gene silencing.89 Overall, promoter methylation of C/EBPα and loss of its expression appears to be one of the most common genetic events in non–small-cell lung cancer and further corroborate that C/EBPα is indeed a tumor suppressor in non–small-cell lung cancer. Moreover, they provide translational opportunities for the reactivation of C/EBPα expression through the use of agents affecting DNA methylation and/or histone acetylation.
THE ROLE OF C/EBPα IN OTHER SOLID TUMORS
C/EBPα is expressed in basal and suprabasal keratinocytes of the epidermis, and forced overexpression of C/EBPα in keratinocytes leads to cell cycle blockade.90,91 C/EBPα participates in the p53-mediated G1 cell cycle arrest in response to ultraviolet B radiation or chemotherapy.92 Its expression is diminished or undetectable in the majority of human and mouse squamous cell carcinomas and is associated with Ras expression.93 Forced expression of oncogenic Ras leads to downregulation of C/EBPα in immortalized keratinocytes, and C/EBPα expression leads to G1 cell cycle arrest in squamous cell carcinoma cell lines.94 These findings suggest that C/EBPα is an important regulator of growth control and DNA damage response in the skin. Epidermal-specific C/EBPα-knockout mice survive and display no detectable abnormalities in epidermal keratinocyte proliferation.95 However, C/EBPα-null mice are highly susceptible to dimethylbezanthrcene-induced skin tumor development. These results provide genetic evidence for the tumor suppressive role of C/EBPα in skin carcinogenesis.
C/EBPα is expressed in normal prostate epithelium with strong expression observed in the basal layer.96 In most prostate adenocarcinomas it is expressed at low levels. Quite surprisingly, overexpression of C/EBPα in two prostate cancer cell lines led to increased proliferation rates, the mechanism of which was not defined.96 Whether these observations are cell-line specific or represent a more general phenomenon in prostate epithelium remains to be seen.
C/EBPα is expressed at high levels in terminally differentiated, mature liver hepatocytes, while it is virtually absent in regenerating liver and hepatomas15 suggesting that C/EBPα expression might be inversely correlated with proliferation. Indeed, overexpression of C/EBPα in hepatocytes leads to impaired proliferation and tumorigenicity in human hepatoma cell lines.97 Liver-specific C/EBPα–knock-in mice demonstrate partial resistance to diethylnitrosamine-induced hepatocellular carcinoma formation corroborating a tumor suppressive role of C/EBPα in hepatic carcinogenesis.98
Downregulation of C/EBPα in squamous cell cancers of the head and neck region has been reported correlating with degree of promoter methylation, while no mutations were found in 59 tumor specimens examined.99 C/EBPα expression in cell lines led to growth inhibition. One microarray study identified downregulation of C/EBPα as a marker for extensive lymph node metastases.100
TARGETING C/EBPα TO FIGHT CANCER
C/EBPα is required for granulocytic differentiation, and restoration of its proper function holds the promise of enabling leukemic stem and progenitor cells from patients with AML and CML myeloid blast crisis to overcome the block of differentiation and undergo maturation into functional effector cells. In mouse models of AML and acute promyelocytic leukemia, overexpression of C/EBPα has resulted in abrogation of the leukemia and in increased survival of the animals.101–103 Of course, replacement therapy for a nuclear transcription factor is a serious technical challenge. Gene-therapy utilizing novel vector systems, such as lentiviral vectors or fusion peptides, such as VP22 or TAT to facilitate entry into the cells are potential considerations.104–107 Compound nanoparticles that target a TAT-bound protein to specific cell subsets via antibody binding to surface markers, as recently described could be exploited to deliver functional C/EBPα to the cytoplasm and nucleus of leukemic blasts via for example a CD33 antibody targeting myeloid cells in order to induce granulocytic-monocytic differentiation.108 This approach seems potentially feasible since virtually all AML blasts express CD33, and a calicheamicin-bound anti-CD33 antibody, gemtuzumab is already in clinical use.109
A more pragmatic approach to restore or activate the C/EBPα differentiation pathway is through small molecule modulators of the expression of C/EBPα, the C/EBPα protein itself or the differentiation pathway governed by C/EBPα. E.g. ATRA upregulates C/EBPα mRNA and protein in myeloid cells.28 ATRA is being used clinically for the treatment of acute promyelocytic leukemia, and recently, it has been employed successfully in AML patients with NPM mutations.110 However, it is not known whether C/EBPα upregulation contributes to the favorable response of these patients to ATRA, as ATRA also greatly increases expression of C/EBPβ which can itself induce maturation of acute promyelocytic leukemia cells.111 C/EBPα translation can be efficiently stimulated in vitro using the triterpenoid antineoplastic drug 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid (CDDO), which increases the translation of p42 CEPBα and the ratio of p42/p30 C/EBPα, leading to granulocytic differentiation in myeloid cells.112 High levels of BCR-ABL, as present in CML blast crisis, induce hnRNP-E2 expression via a MAPK-dependent mechanism. The MEK1/2 inhibitor CI-1040 inhibits hnRNP-E2 expression, and through its association with C/EBPα mRNA, results in increased C/EBPα protein expression and granulocytic differentiation of BCR-ABL–expressing 32D cells.71 MEK inhibitors also inhibit serine-21 phosphorylation of C/EBPα, leading to a more active protein and to enhanced granulocytic differentiation.74 Dephosphorylation of C/EBPα Ser21 was also seen with a Flt3 tyrosine kinase inhibitor,74 and Flt3 kinase inhibitors are the subject of intense clinical study in AML. Our group has also developed luciferase-based assays that are capable of monitoring the modulation of C/EBPα activity in high-throughput assays utilizing a C/EBP-regulated promoter construct. Such assays now allow the screening of millions of small molecules to hopefully lead to the discovery of novel compounds that will indeed restore C/EBPα function and will be lead compounds in further clinical development for the treatment of AML and other cancers where the C/EBPα pathway is involved.
CONCLUSION
Research done over the past several years clearly points out the critical tissue- and cell-specific function of C/EBPα at the crossroads of cell proliferation and differentiation control. Also, evidence now appears overwhelming to document the role of C/EBPα as a bona fide tumor suppressor silenced in multiple different ways in a wide range of leukemias in which its mutational status and transcriptional signature has led to new diagnostic and prognostic classification schemes. The downregulation and inactivation of C/EBPα might be a required step in the development of several other tumor types indicating a wider role for C/EBPα as a tumor suppressor. Better understanding of this prototypical differentiation factor will likely open doors for novel therapeutic strategies to modulate critical differentiation pathways in cancer.
Acknowledgment
We apologize to colleagues whose original work could not be cited due to space limitations.
Footnotes
Supported by National Institutes of Health/National Cancer Institute's “SPORE in Human Lung Cancer” P50 CA90578-04 (B.H. and D.G.T.) and RO1 CA118316 (D.G.T.), the Flight Attendant Medical Research Institute (B.H. and E.L.), the International Association for the Study of Lung Cancer (E.L.), and a grant of the Deutsche Forschungsgemeinschaft (DFG KO2155/1-1 and /2-1; S.K.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Steffen Koschmieder, Balazs Halmos, Daniel G. Tenen
Collection and assembly of data: Steffen Koschmieder, Balazs Halmos, Daniel G. Tenen
Data analysis and interpretation: Steffen Koschmieder, Balazs Halmos, Daniel G. Tenen
Manuscript writing: Steffen Koschmieder, Balazs Halmos, Elena Levantini, Daniel G. Tenen
Final approval of manuscript: Steffen Koschmieder, Balazs Halmos, Elena Levantini, Daniel G. Tenen
REFERENCES
- 1.Tenen DG, Hromas R, Licht JD, et al. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 2.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of C/EBPα, encoding CCAAT/enhancer binding protein-alpha (C/EBP alpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 3.Porse BT, Bryder D, Theilgaard-Monch K, et al. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J Exp Med. 2005;202:85–96. doi: 10.1084/jem.20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 5.Landschulz WH, Johnson PF, Adashi EY, et al. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 6.Akira S, Isshiki H, Sugita T, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. Embo J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roman C, Platero JS, Shuman J, et al. Ig/EBP-1: A ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990;4:1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- 8.Cleutjens CB, van Eekelen CC, van Dekken H, et al. The human C/EBP delta (CRP3/CELF) gene: Structure and chromosomal localization. Genomics. 1993;16:520–523. doi: 10.1006/geno.1993.1220. [DOI] [PubMed] [Google Scholar]
- 9.Antonson P, Stellan B, Yamanaka R, et al. A novel human CCAAT/enhancer binding protein gene, C/EBPepsilon, is expressed in cells of lymphoid and myeloid lineages and is localized on chromosome 14q11.2 close to the T-cell receptor alpha/delta locus. Genomics. 1996;35:30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 10.Park JS, Luethy JD, Wang MG, et al. Isolation, characterization, and chromosomal localization of the human GADD153 gene. Gene. 1992;116:259–267. doi: 10.1016/0378-1119(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 11.Miller M, Shuman JD, Sebastian T, et al. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem. 2003;278:15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 12.Lin FT, MacDougald OA, Diehl AM, et al. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: Transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci U S A. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability cooperate to activate transcription in both yeast and mammalian cells. Embo J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman AD, McKnight SL. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 15.Birkenmeier EH, Gwynn B, Howard S, et al. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 16.Antonson P, Xanthopoulos KG. Molecular cloning, sequence, and expression patterns of the human gene encoding CCAAT/enhancer binding protein alpha (C/EBP alpha) Biochem Biophys Res Commun. 1995;215:106–113. doi: 10.1006/bbrc.1995.2439. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 18.McNagny KM, Sieweke MH, Doderlein G, et al. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. Embo J. 1998;17:3669–3680. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi Y, Nishio H, Kishi K, et al. C/EBPbeta and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: Implication for C/EBP beta activity in eosinophil gene expression. Blood. 1999;94:1429–1439. [PubMed] [Google Scholar]
- 20.Cassel TN, Suske G, Nord M. C/EBP alpha and TTF-1 synergistically transactivate the Clara cell secretory protein gene. Ann N Y Acad Sci. 2000;923:300–302. doi: 10.1111/j.1749-6632.2000.tb05537.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang ND, Finegold MJ, Bradley A, et al. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 22.Sugahara K, Iyama KI, Kimura T, et al. Mice lacking CCAAt/enhancer-binding protein-alpha show hyperproliferation of alveolar type II cells and increased surfactant protein mRNAs. Cell Tissue Res. 2001;306:57–63. doi: 10.1007/s004410100420. [DOI] [PubMed] [Google Scholar]
- 23.Flodby P, Barlow C, Kylefjord H, et al. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 24.Zhang DE, Zhang P, Wang ND, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha–deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Yang J, Croniger CM, Lekstrom-Hines J, et al. Metabolic response of mice to postnatal deletion of CAAT/enhancer-binding protein. J Biol Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- 25.Rosenbauer F, Koschmieder S, Steidl U, et al. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106:1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang DE, Hetherington CJ, Meyers S, et al. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Iwama A, Datta MW, et al. Upregulation of interleukin 6 and granulocyte colony-stimulating factor receptors by transcription factor CCAAT enhancer binding protein alpha (C/EBP alpha) is critical for granulopoiesis. J Exp Med. 1998;188:1173–1184. doi: 10.1084/jem.188.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radomska HS, Huettner CS, Zhang P, et al. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBP alpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Iwasaki-Arai J, Iwasaki H, et al. Enhancement of hematopoietic stem-cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Johansen LM, Iwama A, Lodie TA, et al. C-Myc is a critical target for c/EBP alpha in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang QF, Cleaves R, Kummalue T, et al. Cell cycle inhibition mediated by the outer surface of the C/EBP alpha basic region is required but not sufficient for granulopoiesis. Oncogene. 2003;22:2548–2557. doi: 10.1038/sj.onc.1206360. [DOI] [PubMed] [Google Scholar]
- 33.Timchenko NA, Wilde M, Nakanishi M, et al. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Iakova P, Wilde M, et al. C/EBP alpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen TA, Kowenz-Leutz E, Leutz A, et al. Cooperation between C/EBP alpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slomiany BA, D'Arigo KL, Kelly MM, et al. C/EBP alpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol Cell Biol. 2000;20:5986–5997. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porse BT, Pedersen TA, Xu X, et al. E2F repression by C/EBP alpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–258. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 38.Preudhomme C, Sagot C, Boissel N, et al. Favorable prognostic significance of C/EBPα mutations in patients with de novo acute myeloid leukemia: A study from the Acute Leukemia French Association (ALFA) Blood. 2002;100:2717–2723. doi: 10.1182/blood-2002-03-0990. [DOI] [PubMed] [Google Scholar]
- 39.Gombart AF, Hofmann WK, Kawano S, et al. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood. 2002;99:1332–1340. doi: 10.1182/blood.v99.4.1332. [DOI] [PubMed] [Google Scholar]
- 40.Snaddon J, Smith ML, Neat M, et al. Mutations of C/EBPα in acute myeloid leukemia FAB types M1 and M2. Genes Chromosomes Cancer. 2003;37:72–78. doi: 10.1002/gcc.10185. [DOI] [PubMed] [Google Scholar]
- 41.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, et al. Biallelic mutations in the C/EBPα gene and low C/EBPα expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4:31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 42.Fröhling S, Schlenk RF, Stolze I, et al. C/EBPα mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–633. doi: 10.1200/JCO.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 43.Fröhling S, Schlenk RF, Krauter J, et al. Acute myeloid leukemia with deletion 9q within a noncomplex karyotype is associated with C/EBPα loss-of-function mutations. Genes Chromosomes Cancer. 2005;42:427–432. doi: 10.1002/gcc.20152. [DOI] [PubMed] [Google Scholar]
- 44.Smith ML, Cavenagh JD, Lister TA, et al. Mutation of C/EBPα in familial acute myeloid leukemia. N Engl J Med. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 45.Bienz M, Ludwig M, Leibundgut EO, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 46.Shih LY, Huang CF, Lin TL, et al. Heterogeneous patterns of CEBP alpha mutation status in the progression of myelodysplastic syndrome and chronic myelomonocytic leukemia to acute myelogenous leukemia. Clin Cancer Res. 2005;11:1821–1826. doi: 10.1158/1078-0432.CCR-04-1932. [DOI] [PubMed] [Google Scholar]
- 47.Lin LI, Chen CY, Lin DT, et al. Characterization of C/EBPα mutations in acute myeloid leukemia: Most patients with C/EBPα mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11:1372–1379. doi: 10.1158/1078-0432.CCR-04-1816. [DOI] [PubMed] [Google Scholar]
- 48.Wouters BJ, Jorda MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced C/EBPα and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 50.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 51.Bacher U, Haferlach C, Kern W, et al. Prognostic relevance of FLT3-TKD mutations in AML: The combination matters—An analysis of 3,082 patients. Blood. 2008;111:2527–2537. doi: 10.1182/blood-2007-05-091215. [DOI] [PubMed] [Google Scholar]
- 52.Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–6837. doi: 10.1038/sj.onc.1210765. [DOI] [PubMed] [Google Scholar]
- 53.Nerlov C. C/EBP alpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 54.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBP alpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 55.Wagner K, Zhang P, Rosenbauer F, et al. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc Natl Acad Sci U S A. 2006;103:6338–6343. doi: 10.1073/pnas.0508143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 57.Sellick GS, Spendlove HE, Catovsky D, et al. Further evidence that germline C/EBPα mutations cause dominant inheritance of acute myeloid leukaemia. Leukemia. 2005;19:1276–1278. doi: 10.1038/sj.leu.2403788. [DOI] [PubMed] [Google Scholar]
- 58.Nanri T, Uike N, Kawakita T, et al. A pedigree harboring a germ-line N-terminal C/EBP mutation and development of acute myeloblastic leukemia with a somatic C-Terminal C/EBP mutation. Blood. 2006;108(suppl) abstr 543. [Google Scholar]
- 59.Chapiro E, Russell L, Radford-Weiss I, et al. Overexpression of C/EBPα resulting from the translocation t(14;19)(q32;q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;108:3560–3563. doi: 10.1182/blood-2006-03-010835. [DOI] [PubMed] [Google Scholar]
- 60.Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109:3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 61.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 62.Fitzgibbon J, Smith LL, Raghavan M, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res. 2005;65:9152–9154. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 63.Raghavan M, Lillington DM, Skoulakis S, et al. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–378. [PubMed] [Google Scholar]
- 64.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 65.Chim CS, Liang R, Kwong YL. Hypermethylation of gene promoters in hematological neoplasia. Hematol Oncol. 2002;20:167–176. doi: 10.1002/hon.694. [DOI] [PubMed] [Google Scholar]
- 66.Chim CS, Wong AS, Kwong YL. Infrequent hypermethylation of C/EBPα promotor in acute myeloid leukaemia. Br J Haematol. 2002;119:988–990. doi: 10.1046/j.1365-2141.2002.03952.x. [DOI] [PubMed] [Google Scholar]
- 67.Lin TC, Lee CY, Tien HF, et al. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in acute myeloid leukemia. Blood. 2007;110(suppl) abstr 2113. [Google Scholar]
- 68.Keeshan K, He Y, Wouters BJ, et al. Tribbles homolog 2 inactivates C/EBP alpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10:401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hackanson B, Bennett KL, Brena RM, et al. Dual epigenetic control of CCAAT/enhancer binding protein (C/EBP) expression in acute myeloid leukemia. Blood. 2007;110(suppl) abstr 2116. [Google Scholar]
- 70.Perrotti D, Cesi V, Trotta R, et al. BCR-ABL suppresses C/EBP alpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 71.Chang JS, Santhanam R, Trotta R, et al. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBP alpha-driven myeloid differentiation. Blood. 2007;110:994–1003. doi: 10.1182/blood-2007-03-078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Helbling D, Mueller BU, Timchenko NA, et al. The leukemic fusion gene AML1-MDS1-EVI1 suppresses C/EBPα in acute myeloid leukemia by activation of Calreticulin. Proc Natl Acad Sci U S A. 2004;101:13312–13317. doi: 10.1073/pnas.0404731101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helbling D, Mueller BU, Timchenko NA, et al. CBFB-SMMHC is correlated with increased calreticulin expression and suppresses the granulocytic differentiation factor C/EBPα in AML with inv16. Blood. 2005;106:1369–1375. doi: 10.1182/blood-2004-11-4392. [DOI] [PubMed] [Google Scholar]
- 74.Radomska HS, Basseres DS, Zheng R, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203:371–381. doi: 10.1084/jem.20052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tremblay M, Girard N, Haman A, et al. The RARa-PLZF oncogenic protein inhibits C/EBPa function in myeloid cells. Blood. 2007;110(suppl) abstr 1825. [Google Scholar]
- 76.Trivedi AK, Bararia D, Christopeit M, et al. Proteomic identification of C/EBP-DBD multiprotein complex: JNK1 activates stem cell regulator C/EBP alpha by inhibiting its ubiquitination. Oncogene. 2007;26:1789–1801. doi: 10.1038/sj.onc.1209964. [DOI] [PubMed] [Google Scholar]
- 77.Geletu M, Balkhi MY, Peer Zada AA, et al. Target proteins of C/EBP alpha p30 in AML: C/EBP alpha p30 enhances sumoylation of C/EBP alpha p42 via up-regulation of Ubc9. Blood. 2007;110:3301–3309. doi: 10.1182/blood-2007-01-071035. [DOI] [PubMed] [Google Scholar]
- 78.Whitsett JA, Matsuzaki Y. Transcriptional regulation of perinatal lung maturation. Pediatr Clin North Am. 2006;53:873–887. viii. doi: 10.1016/j.pcl.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Cassel TN, Berg T, Suske G, et al. Synergistic transactivation of the differentiation-dependent lung gene Clara cell secretory protein (secretoglobin 1a1) by the basic region leucine zipper factor CCAAT/enhancer-binding protein alpha and the homeodomain factor Nkx2.1/thyroid transcription factor-1. J Biol Chem. 2002;277:36970–36977. doi: 10.1074/jbc.M201293200. [DOI] [PubMed] [Google Scholar]
- 80.Cassel TN, Nordlund-Moller L, Andersson O, et al. C/EBP alpha and C/EBP delta activate the Clara cell secretory protein gene through interaction with two adjacent C/EBP-binding sites. Am J Respir Cell Mol Biol. 2000;22:469–480. doi: 10.1165/ajrcmb.22.4.3916. [DOI] [PubMed] [Google Scholar]
- 81.Nord M, Cassel TN, Braun H, et al. Regulation of the Clara cell secretory protein/uteroglobin promoter in lung. Ann N Y Acad Sci. 2000;923:154–165. doi: 10.1111/j.1749-6632.2000.tb05527.x. [DOI] [PubMed] [Google Scholar]
- 82.Bassères DS, Levantini E, Ji H, et al. Respiratory failure due to differentiation arrest and expansion of alveolar cells following lung-specific loss of the transcription factor C/EBP alpha in mice. Mol Cell Biol. 2006;26:1109–1123. doi: 10.1128/MCB.26.3.1109-1123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martis PC, Whitsett JA, Xu Y, et al. C/EBP alpha is required for lung maturation at birth. Development. 2006;133:1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- 84.Halmos B, Huettner CS, Kocher O, et al. Down-regulation and antiproliferative role of C/EBP alpha in lung cancer. Cancer Res. 2002;62:528–534. [PubMed] [Google Scholar]
- 85.Costa DB, Li S, Kocher O, et al. Immunohistochemical analysis of C/EBP alpha in non–small-cell lung cancer reveals frequent down-regulation in stage II and IIIA tumors: A correlative study of E3590. Lung Cancer. 2007;56:97–103. doi: 10.1016/j.lungcan.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Halmos B, Basseres DS, Monti S, et al. A transcriptional profiling study of CCAAT/enhancer binding protein targets identifies hepatocyte nuclear factor 3 beta as a novel tumor suppressor in lung cancer. Cancer Res. 2004;64:4137–4147. doi: 10.1158/0008-5472.CAN-03-4052. [DOI] [PubMed] [Google Scholar]
- 87.Girard L, Zochbauer-Muller S, Virmani AK, et al. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small-cell lung cancer and non–small-cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 88.Yoshida Y, Hughes DE, Rausa FM, 3rd, et al. C/EBP alpha and HNF6 protein complex formation stimulates HNF6-dependent transcription by CBP coactivator recruitment in HepG2 cells. Hepatology. 2006;43:276–286. doi: 10.1002/hep.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tada Y, Brena RM, Hackanson B, et al. Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer. J Natl Cancer Inst. 2006;98:396–406. doi: 10.1093/jnci/djj093. [DOI] [PubMed] [Google Scholar]
- 90.Maytin EV, Habener JF. Transcription factors C/EBP alpha, C/EBP beta, and CHOP (Gadd153) expressed during the differentiation program of keratinocytes in vitro and in vivo. J Invest Dermatol. 1998;110:238–246. doi: 10.1046/j.1523-1747.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- 91.Zhu S, Yoon K, Sterneck E, et al. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoon K, Smart RC. C/EBP alpha is a DNA damage–inducible p53-regulated mediator of the G1 checkpoint in keratinocytes. Mol Cell Biol. 2004;24:10650–10660. doi: 10.1128/MCB.24.24.10650-10660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh HS, Smart RC. Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J Invest Dermatol. 1998;110:939–945. doi: 10.1046/j.1523-1747.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 94.Shim M, Powers KL, Ewing SJ, et al. Diminished expression of C/EBP alpha in skin carcinomas is linked to oncogenic Ras and reexpression of C/EBP alpha in carcinoma cells inhibits proliferation. Cancer Res. 2005;65:861–867. [PubMed] [Google Scholar]
- 95.Loomis KD, Zhu S, Yoon K, et al. Genetic ablation of CCAAT/enhancer binding protein alpha in epidermis reveals its role in suppression of epithelial tumorigenesis. Cancer Res. 2007;67:6768–6776. doi: 10.1158/0008-5472.CAN-07-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yin H, Radomska HS, Tenen DG, et al. Down regulation of PSA by C/EBP alpha is associated with loss of AR expression and inhibition of PSA promoter activity in the LNCaP cell line. BMC Cancer. 2006;6:158. doi: 10.1186/1471-2407-6-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Watkins PJ, Condreay JP, Huber BE, et al. Impaired proliferation and tumorigenicity induced by CCAAT/enhancer-binding protein. Cancer Res. 1996;56:1063–1067. [PubMed] [Google Scholar]
- 98.Tan EH, Hooi SC, Laban M, et al. CCAAT/enhancer binding protein alpha knock-in mice exhibit early liver glycogen storage and reduced susceptibility to hepatocellular carcinoma. Cancer Res. 2005;65:10330–10337. doi: 10.1158/0008-5472.CAN-04-4486. [DOI] [PubMed] [Google Scholar]
- 99.Bennett KL, Hackanson B, Smith LT, et al. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67:4657–4664. doi: 10.1158/0008-5472.CAN-06-4793. [DOI] [PubMed] [Google Scholar]
- 100.Roepman P, Wessels LF, Kettelarij N, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37:182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 101.Truong BT, Lee YJ, Lodie TA, et al. CCAAT/Enhancer binding proteins repress the leukemic phenotype of acute myeloid leukemia. Blood. 2003;101:1141–1148. doi: 10.1182/blood-2002-05-1374. [DOI] [PubMed] [Google Scholar]
- 102.Lee YJ, Jones LC, Timchenko NA, et al. CCAAT/enhancer binding proteins alpha and epsilon cooperate with all-trans-retinoic acid in therapy but differ in their antileukemic activities. Blood. 2006;108:2416–2419. doi: 10.1182/blood-2006-02-003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ferrari-Amorotti G, Keeshan K, Zattoni M, et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBP alpha. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miyoshi H, Smith KA, Mosier DE, et al. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 105.Elliott G, O'Hare P, editors. Intercellular trafficking and protein delivery by a herpes virus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 106.Schwarze SR, Ho A, Vocero-Akbani A, et al. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 107.Zhou Y, Du W, Koretsky T, et al. TAT-mediated intracellular delivery of NPM-derived peptide induces apoptosis in leukemic cells and suppresses leukemogenesis in mice. Blood. 2008;112:2474–2483. doi: 10.1182/blood-2007-12-130211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu X, Wang Y, Nakamura K, et al. Cell studies of a three-component antisense MORF/tat/Herceptin nanoparticle designed for improved tumor delivery. Cancer Gene Ther. 2008;15:126–132. doi: 10.1038/sj.cgt.7701111. [DOI] [PubMed] [Google Scholar]
- 109.Bross PF, Beitz J, Chen G, et al. Approval summary: Gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res. 2001;7:1490–1496. [PubMed] [Google Scholar]
- 110.Schlenk RF, Döhner K, Kneba M, et al. The genotype NPM1mut/FLT3-ITDneg is a highly significant predictive factor for response to therapy with all-trans retinoic acid in acute myeloid leukemia: Results from AMLSG trial AML HD98B. Blood. 2007;110:297A. [Google Scholar]
- 111.Duprez E, Wagner K, Koch H, et al. C/EBPbeta: A major PML-RARA-responsive gene in retinoic acid-induced differentiation of APL cells. Embo J. 2003;22:5806–5816. doi: 10.1093/emboj/cdg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koschmieder S, D'Alo F, Radomska H, et al. CDDO induces granulocytic differentiation of myeloid leukemic blasts through translational up-regulation of p42 CCAAT enhancer binding protein alpha. Blood. 2007;110:3695–3705. doi: 10.1182/blood-2006-11-058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Akashi K, Traver D, Miyamoto T, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 114.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBP alpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]