The development of cryo-EM into a mainstream structural biology technique
. Author manuscript; available in PMC: 2016 Jul 1.
Published in final edited form as: Nat Methods. 2016 Jan;13(1):24–27. doi: 10.1038/nmeth.3694
Abstract
Single-particle cryo-electron microscopy (cryo-EM) has emerged over the last two decades as a technique capable of studying challenging systems that otherwise defy structural characterization. Recent technical advances have resulted in a ‘quantum leap’ in applicability, throughput and achievable resolution that has gained this technique worldwide attention. Here I discuss some of the major historical landmarks in the development of the cryo-EM field, ultimately leading to its present success.
A unique structural biology niche
Structural biology techniques have been developed to visualize the architecture of macromolecular assemblies and have proven invaluable to our mechanistic understanding of biomolecular function. To date, the most broadly used method to obtain atomic-detail macromolecular structures is X-ray crystallography. However, this approach is limited by the requirement for large amounts of sample and by the bottleneck of protein crystallization, which is often far from straightforward. Although NMR does not require crystallization, it also requires large sample amounts as well as isotopic enrichment, and size limitations have generally restricted it to small proteins, small RNAs or single domains. The constraints of these ‘classical’ structural techniques have imposed limitations in their application to large complexes, integral membrane proteins, polymers and macromolecular assemblies for which multiple conformational or compositional states coexist.
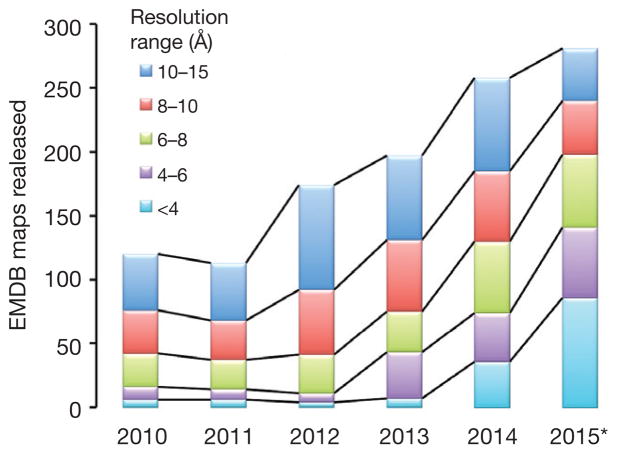
Single-particle cryo-electron microscopy (cryo-EM) has emerged over the past two decades as a structural biology technique applicable to the study of challenging biological systems because it does not entail crystallization and requires only small amounts of sample. Recently, advances in instrumentation and software have dramatically improved the potential of singleparticle cryo-EM to produce structures at resolutions that allow direct atomic modeling into the density map as well as to deal with compositional and conformational mixtures. Furthermore, the time needed to solve a structure has greatly diminished as a result of automation in both data collection and processing and of the requirement for fewer particle images to build the signal required for high resolution (thanks to new detector technology; see Box 1). Databases are now being flooded by new cryo-EM structure depositions at an unprecedented rate (Fig. 1), including those for critical macromolecular assemblies that until very recently were only pie-in-the-sky structural goals. Thus, we are experiencing a quantum leap in the applicability, throughput and achievable resolution of single-particle cryo-EM that has made this technique gain worldwide attention.
BOX 1. THE DETECTION FACTOR.
Traditionally, EM images have been recorded on film, which needs to be developed and then digitized. The advent of CCD (charge-coupled device) cameras allowed the automation of the data-collection process and thus the growth of data sets in both number and size. However, a limitation with CCDs stems from the fact that electrons must first be converted to photons via interaction with a scintillator. Multiple scattering events of the electrons in the scintillator give rise to a broad ‘cloud’ of photons, much larger than the CCD pixels, for each electron contributing to the image. The result is a severe attenuation in signal at high resolution, a big problem for an imaging method that is limited by signal to start with.
One way around these limitations is to detect electrons directly on a silicon wafer. To realize this approach, a number of engineering hurdles needed to be overcome, chief among them the development of a radiation-hardened design that would reduce damage to the silicon chip and thus allow the use of the detector for a practical number of exposures. Multiple efforts over the years, one in the UK (led by Richard Henderson, Greg McMullan and Wasi Faruqi at the Medical Research Council Laboratory of Molecular Biology) and two in the United States (led by Peter Denes at Lawrence Berkeley National Laboratory and Nguyen-Huu Xuong at the University of California, San Diego), resulted in engineering solutions that were ultimately commercialized by three different companies. These direct electron-detection cameras (or direct detectors) have little or no noise and result in high contrast and preservation of high-resolution signal. Additionally, they come with the added bonus of very fast readouts. This last property is what has in recent years allowed—as pioneered by Niko Grigorieff and colleagues27—the collection of cryo-EM data in ‘movie mode’ for correcting beam-induced motion. Before correction, most images are blurred and thus lack high-resolution information. Quite typically, one image out of 20 or even 50 would be free of this blurring, making high-quality data collection highly inefficient. The new cameras today allow a relatively simple correction process, such that close to 100% of the images contain high-resolution data. These cameras represent the hardware component of the ‘quantum leap’ in single-particle cryo-EM we are experiencing today.
Figure 1.
Increase in the number of EM map depositions in the Electron Microscopy Data Bank (http://emdatabank.org/) over the last six years. Asterisk indicates that data for 2015 are incomplete (through 14 October). The most notable growth is in the number of structures at better than 4-Å resolution in the last two years. (Courtesy of C. Lawson.)
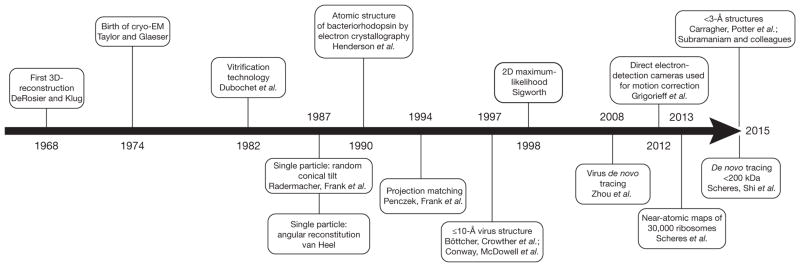
Here I give my personal perspective on the major historical steps in the development of the three-dimensional (3D) reconstruction EM field, ultimately leading to the present-day success of this technique (Fig. 2). This short Commentary is not, however, intended as a comprehensive account of all the many contributions, made by many labs, to the progress of the field.
Figure 2.
Timeline of key contributions to the field of 3D electron microscopy highlighted in this Commentary.
The development of 3D cryo-EM
Biological samples can be directly visualized at the molecular level using transmission electron microscopy (TEM). TEM images produce 2D projections of the object, and projections from different directions (views) can be computationally combined to produce a 3D reconstruction. In the late 1960s, David DeRosier and Aaron Klug used Fourier-Bessel principles to combine multiple views of the molecules naturally present in the helical arrangement of the T4 bacteriophage tail1. And so 3D-EM reconstruction was born.
Three major methodological bottlenecks needed to be overcome, however, to establish the technique as a general method. First, the sample needs to be preserved under the ultra-high-vacuum conditions of the TEM. A solution was also needed to overcome the radiation damage caused by high-energy electrons hitting the sample. And finally, a mechanism needed to be found to overcome the fact that biological samples, which are made of light elements that scatter electrons similarly to the water background, show very low contrast.
The first practical solution to these difficulties, applied by DeRosier and Klug and still in practice for certain samples, was the use of heavy-atom salts, such as uranyl acetate, as staining reagents to generate a dried ‘cast’ of the molecules before insertion in the scope. Heavy-metal staining reduces the problem of radiation damage, as it is the inorganic cast, not the protein itself, that is imaged. This method also generates a high-contrast image due to the high atomic number of uranium (because the proteins scatters less that the surrounding stain, the contrast appears reversed and this method is known as ‘negative staining’). However, staining and drying the samples comes at a cost, as the stain may not be homogeneous or penetrate small cavities in the sample, and drying can potentially collapse the structures. Most importantly, staining limits the resolution because of the grain size of the heavy atom salt (to ~15 Å in the most optimal cases).
The breakthrough that opened the door to high-resolution structure determination came from the demonstration by Ken Taylor and Robert Glaeser2 that cryoprotection of frozen-hydrated samples allowed the preservation of high resolution features in proteins. This work initiated the field of cryo-EM. Later work from Dubochet and colleagues3 resulted in the development of a practical method for vitrification of samples (by cryo-plunging of thin solutions into a potent cryogen such as liquid ethane or liquid propane). Together with the commercialization of stable cryo-stages, these advances opened the door to the broad use of cryo-EM and allowed the field to expand.
Helical samples, viruses and 2D crystals were favorite specimens as 3D reconstruction methodology was being developed. Of special mention is the pioneering work by Richard Henderson and colleagues on 2D electron crystallography, ultimately leading to the atomic structure of bacteriorhodopsin4 that was used for homology modeling of seven-helix G protein–coupled receptors for many years. Henderson and colleagues used a combination of images and electron diffraction patterns from 2D crystals. The challenges faced by this technique include the need to tilt the sample in order to obtain different views of the molecule and the requirement of generating perfectly flat 2D crystals (a difficult task, as they are typically only one protein thick!). As a result, only a handful of structures have been pursued by electron crystallography, and only a fraction ever reached atomic resolution. Nevertheless, electron crystallography has produced a number of atomic models of high biological relevance, including those of the plant light-harvesting complex5 and tubulin6, and culminating with the highest-resolution structure obtained by 3D-EM to date, that of aquaporin 0 at 1.9 Å by Tom Walz and Steve Harrison7. A benefit of electron crystallography is that it can be used to study integral membrane proteins in their native membrane environment, as was demonstrated for both bacteriorhodopsin and aquaporin 0.
Some well-ordered helical arrangements, which can be regarded as being 1D crystals (either naturally occurring or induced), were also able to reach close to atomic resolution by cryo-EM. Among them is the acetylcholine receptor studied by Unwin, whose work included a time-resolved study that used a fast freezing process to trap the open state of the channel before inactivation8. Icosahedral viruses, with their inherent advantage of high symmetry for both alignment and boosting of the signal of the final reconstruction, were the first noncrystalline samples to break the nanometer barrier to allow visualization of protein secondary structure9,10 and ultimately to produce atomic-resolution maps11 that allowed atomic modeling of totally new structures directly into the density12.
However, to make 3D-EM a broadly applicable technique, new algorithms and reconstruction strategies were needed for macromolecules not organized into icosahedral, helical or crystalline arrays. Methodology to deal with samples made of individual macromolecular complexes, ‘single particles’, that adopt random (or at least multiple) orientations on the EM grid was developed in the pioneering work of Joachim Frank and co-workers13. Their efforts led to the birth and growth of single-particle reconstruction, the general technique in broad use today for cryo-EM–based structure determination.
Challenges and opportunities
The key to single-particle cryo-EM was the development of computational tools to define the relative orientations of the 2D projection images in order to produce a 3D reconstruction. The two major challenges in this endeavor concern the noisy character of the images and the need to computationally identify their relative orientations. In order to minimize radiation damage to the sample, very low electron doses are used during cryo-EM data collection (~20–40 electrons/Å2). To gain signal, tens to hundreds of thousands of images of single particles need to be averaged (and before the new detector technology was available, this number was in the millions). Several computational methods have been developed to identify the relative orientations of the single-particle projection images.
One of these methods is the “projection matching” approach developed by Frank and Pawel Penczek14. In projection matching, each experimental particle image is compared with computationally generated views of a 3D reference that resembles the true structure, and angles are assigned based on a cross-correlation criterion. Initial reference structures may need to be determined de novo from EM images, using geometry-based approaches (in which two or more images of the same area of the specimen at different tilt angles are collected) that are generally considered robust and that provide information on the handedness of the structure. Among such approaches is the random conical tilt reconstruction method developed by Frank and Michael Radermacher15. As an alternative to geometry-based methods, the relative orientation between particle images can be analytically determined using the fact that each pair of 2D projections shares a ‘common line’ in the 3D Fourier transform16. The popular angular reconstitution method developed by Marin van Heel17 is a real-space implementation of this common lines principle. This method does not require specimen tilting, but it does not define the handedness of the object.
These different image-analysis methodologies have been implemented in software packages that are broadly used by the EM community, such as Spider18 and Imagic19. EMAN20 and latter EMAN2 (ref. 21) were developed by Wah Chiu and Steve Ludtke with the purpose of making image processing more easily accessible to non-experts. Frealign22, developed by Niko Grigorieff, focuses on 3D refinement (the process by which the reference density is iteratively improved together with the assignment of relative angles for the experimental images, leading to improve resolution) and incorporated a very successful correction of the contrast transfer function of the microscope (the effect of the optical system on the image as a function of spatial frequency) into the reconstruction procedure. SPARX23 was created by Penczek, Glaeser, Paul Adams and Ludtke as an environment for end users, both in X-ray crystallography and in cryo-EM, using a graphical programming approach. Xmipp24, developed by Jose María Carazo, Sjors Scheres and colleagues, used an X-windows interface and was created with an emphasis on modularity of image-analysis protocols and the description of structural heterogeneity.
Because cryo-EM is a single-particle technique, cryo-EM samples often present the additional challenge of conformational or compositional heterogeneity, a major reason why high resolution may not be easily achieved. Analysis of heterogeneous samples requires classification of the particle images into structurally homogeneous subsets. This task can be done using geometry-based principles initially, followed by a projection-matching approach using multiple references. A major advance in cryo-EM image analysis was the implementation of maximum-likelihood (ML) approaches, first introduced by Fred Sigworth25. In this ML implementation, an experimental image is assigned not a single relative orientation based on a similarity criterion, but rather a set of probabilities to be in any orientation. Such methods have been demonstrated to robustly handle noise-ridden cryo-EM images. ML approaches for unsupervised classification of particle images have since been implemented to simultaneously determine coexisting states in the sample. Though computationally demanding, the ability to identify and characterize multiple structural states provides invaluable insight into macromolecular dynamics. Today, packages such as Frealign and Xmipp incorporate ML principles. RELION26, a software tool developed by Scheres, has gained worldwide attention for its capacity to deal with heterogeneous samples and for its very user-friendly interface that has made single-particle image analysis accessible to a broader user community.
A new era: more, better and faster
The 3D-EM field has evolved very dramatically since its first application to the T4 bacteriophage tail1. New instrumentation and image-processing methods have been developed, and throughput has increased with the advent of automation in data collection and analysis. In the decade between the first reported cryo-EM virus structures breaking the nanometer barrier and the first de novo tracing of a viral capsid, the field strengthened, the number of labs using cryo-EM grew and the achievable resolution was systematically improved. But the advances have never been so transformative as during the last few years, with the commercialization and use of direct electron-detection cameras (Box 1).
The new detector technology results in images with notably higher contrast, which, together with their fast read-out, allow for a ‘movie mode’ of data collection that significantly overcomes the limitation of ‘beam-induced motion’. As electrons go through the vitrified sample, they induce movement in the ice/particles while the image is being collected, resulting in blurring. An effective correction of this phenomenon is obtained by splitting the total dose into short frames (for example, 20, with typically 1 e/Å2 dose each) and then applying computational frame alignment that minimizes the blurring caused by beam-induced movement, a procedure pioneered by Grigorieff27. The particle images, having higher signal-to-noise ratios, can be aligned more accurately and therefore contribute to higher resolutions, potentially atomic28,29. The improvement in detector technology has not only led to higher-resolution 3D structure determination using smaller data sets30 but also lowered the size limit of the samples that can be visualized in a frozen, hydrated state so that smaller complexes or individual proteins can be studied31,32. Perhaps even more significantly, it also enables better classification of heterogeneous samples, which has led to the identification and description of even rare states33.
Thus, as a result of improvements in both hardware and software, the applicability, resolution and throughput of single-particle cryo-EM have increased dramatically, making it today a powerful alternative to X-ray crystallography. As a consequence, a flood of new biological insights is being gained into systems traditionally refractory to structural characterization. These systems include a growing number of integral membrane proteins34,35; biological polymers such as F-actin36 and microtubules37; large assemblies that can be produced only in very small amounts, such as transcription initiation complexes38; and rare states within a complex biochemical mixture, as seen in studies of eukaryotic translation initiation complexes33,39.
Concluding thoughts
The field of cryo-EM has now attained a level of development such that certain type of samples, like viruses or ribosomes, are now only rarely pursued using X-ray crystallography. A similar shift is starting to occur for integral membrane proteins. As smaller molecular sizes become feasible, the number of systems pursued by cryo-EM will naturally grow, carving out experimental space that before was the realm of X-ray studies. But for this author the excitement is more in the ‘large’ than in the ‘small’. Large-sized macromolecular assemblies, especially when reconstitution becomes limiting and endogenous sources are scarce, have for some time been the domain of cryo-EM. These assemblies tend to be flexible, and although classification methods have come a long way when applied to biochemical mixtures or well-defined conformational states33,39, the continuous motions seen for certain samples40 will challenge classification schemes and set a limit to achievable resolution, at least for a number of years. Still, such studies will be able to reach subnanometer resolution while describing multiple states (or even conformational trajectories). For such cases structural insight from cryo-EM will be most valuable, but even more so if X-ray crystal structures of components can be docked using hybrid methodologies to generate pseudoatomic models. In this context, computational methods for predicting protein structure or interactions will also be able to play an important role going forward.
Both significant technical issues and new opportunities await the field of cryo-EM (see Commentary from Glaeser in this issue). Phase-plate technology, even better detectors, improved and reproducible cryo-EM sample preparation, and new software that can deal with continuous forms of structural variability are all under development by the cryo-EM community and may lead to even further improvements in the near future. Among the immediate challenges we face are the high costs of purchasing and maintaining expensive EM equipment and the enormous computational loads, calling for a new funding model of shared instruments and supercomputer resources. Finally, the growing field of cryo-EM has a clear need for easy-to-use tools for structure validation that can become broadly used by its community.
Acknowledgments
I would like to thank R. Glaeser for his feedback while writing this commentary. The author is supported by grants from the US National Institute of General Medical Sciences and is a Howard Hughes Medical Institute Investigator.
Footnotes
COMPETING FINANCIAL INTERESTS
The author declares no competing financial interests.
References
- 1.De Rosier DJ, Klug A. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 2.Taylor KA, Glaeser RM. Science. 1974;186:1036–1037. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 3.Dubochet J, Lepault J, Freeman R, Berriman JA, Homo JC. J Microsc. 1982;128:219–237. [Google Scholar]
- 4.Henderson R, et al. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 5.Kühlbrandt W, Wang DN. Nature. 1991;350:130–134. doi: 10.1038/350130a0. [DOI] [PubMed] [Google Scholar]
- 6.Nogales E, Wolf SG, Downing KH. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 7.Gonen T, et al. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unwin N. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 9.Böttcher B, Wynne SA, Crowther RA. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 10.Conway JF, et al. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Jin L, Zhou ZH. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press; San Diego: 1996. [Google Scholar]
- 14.Penczek PA, Grassucci RA, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 15.Radermacher M, Wagenknecht T, Verschoor A, Frank J. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 16.Crowther RA. Phil Trans R Soc Lond B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 17.Van Heel M. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 18.Frank J, et al. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 19.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 20.Ludtke SJ, Baldwin PR, Chiu W. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 21.Tang G, et al. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Grigorieff N. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Hohn M, et al. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Scheres SH, Núñez-Ramírez R, Sorzano CO, Carazo JM, Marabini R. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigworth FJ. J Struct Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- 26.Scheres SH. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brilot AF, et al. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. eLife. 2015;4:e06380. doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartesaghi A, et al. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai XC, Fernandez IS, McMullan G, Scheres SHW. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao M, Cao E, Julius D, Cheng Y. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai XC, et al. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernández IS, et al. Science. 2013;342:1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hite RK, et al. Nature. 2015;527:198–203. doi: 10.1038/nature14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du J, Lü W, Wu S, Cheng Y, Gouaux E. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von der Ecken J, et al. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang R, Alushin GM, Brown A, Nogales E. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Fang J, Taatjes DJ, Nogales E. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.des Georges A, et al. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cianfrocco MA, et al. Cell. 2013;152:120–131. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]