An integrated analysis of genes and functional pathways for aggression in human and rodent models
Abstract
Human genome-wide association studies (GWAS), transcriptome analyses of animal models, and candidate gene studies have advanced our understanding of the genetic architecture of aggressive behaviors. However, each of these methods presents unique limitations. To generate a more confident and comprehensive view of the complex genetics underlying aggression, we undertook an integrated, cross-species approach. We focused on human and rodent models to derive eight gene lists from three main categories of genetic evidence: two sets of genes identified in GWAS studies, four sets implicated by transcriptome-wide studies of rodent models, and two sets of genes with causal evidence from online Mendelian inheritance in man (OMIM) and knockout (KO) mice reports. These gene sets were evaluated for overlap and pathway enrichment to extract their similarities and differences. We identified enriched common pathways such as the G-protein coupled receptor (GPCR) signaling pathway, axon guidance, reelin signaling in neurons, and ERK/MAPK signaling. Also, individual genes were ranked based on their cumulative weights to quantify their importance as risk factors for aggressive behavior, which resulted in 40 top-ranked and highly interconnected genes. The results of our cross-species and integrated approach provide insights into the genetic etiology of aggression.
Subject terms: Psychiatric disorders, Genetics
Introduction
Aggressive behavior is an evolutionarily conserved, heritable trait that is essential for survival and fitness. In humans, aggressive behavior is also shaped by societal and cultural constraints. Context-inappropriate aggression can cause great harm to society, families, and individuals, and has been associated with neuropsychiatric disorders, such as attention-deficit/hyperactivity disorder (ADHD) [1, 2], schizophrenia (SCZ), and bipolar disorder (BIP) [3–5].
The heritability of human aggression has been estimated around 50% [6]. Its complex genetic architecture interacts with environmental factors [7–9]. Decades of animal studies have yielded strong neurochemical and physiological evidence that points to conserved common pathways across species such as serotoninergic and dopaminergic neurotransmission and hormonal signaling [10]. These data helped to inform candidate gene selection for human studies on serotonergic and dopaminergic genes (MAOA, 5HTT, HTR1B, HTR2A, DAT, DRD2, DRD4), as well as hormone-related genes (AR, ESR1, AVP, OXTR) [11]. However, the largest meta-analysis of candidate genes performed to date [12] did not find any significant association. Genome-wide association studies (GWAS) of aggression have been underpowered to detect common variants of small penetrance associated with complex phenotypes [11, 12]. The largest GWAS, which was performed by the Early Genetics and Lifecourse Epidemiology (EAGLE) consortium (http://research.lunenfeld.ca/eagle/), reported association with one SNP, rs11126630, at a suggestive significance level (p = 5.3e−08). Among the “classical” candidates evaluated in the EAGLE dataset, only one gene, AVPR1A, encoding the arginin vasopressin receptor 1A, showed a nominal association with aggression (p = 1.6e−03) [13].
Studies of rare human genetic conditions [14] and gene knockouts (KOs) in mice [15, 16] show that many genetic determinants play critical roles in shaping the emotional circuitry of the brain and modulating aggressive behavior [17, 18]. For example, rare mutations in the gene encoding methyl-CpG-binding protein 2 (MeCP2), a chromatin-associated protein involved in transcription regulation, cause Rett syndrome, mental retardation, and increased aggression [19]. Mecp2 KO mice also show increased aggression [20]. Another example is the gene for prion protein, the mutation of which causes inherited prion diseases and aggressive behavior in humans [21]. Mice with depleted prion protein showed increased aggressiveness [22], possibly related to the role of the prion protein in regulating cytoskeleton and associated proteins [23]. Despite convincing evidence supporting these high-risk genes from human single-gene disorders and KO mice studies, none have reached genome-wide significance in GWAS of aggression. The best p-value was for gene LRRC7 in a study of children (p = 4e−06) [24]. LRRC7 interacts with cytoskeleton molecules and is involved in synaptic spine structure and patterning [25]. Lrrc7 KO mice show significantly increased fighting among littermates [26]. Nevertheless, similar pathways such as synaptic development, axon guidance, and MAPK signaling emerge when examining genes from the top GWAS findings (p ≤ 5e−05, ref. [11]). These pathways were also enriched in genes identified through transcriptomic studies of animal models of aggression in mice [27, 28], rats [29], zebrafish [30], and Drosophila [31]. With increasing sample sizes, some of these genes and, perhaps, additional risk loci may emerge in future GWAS.
Meanwhile, we postulate that a cross-species and integrated approach combining different modalities of genetic data can yield a clearer understanding of the genetics of aggression. Our study focuses on several categories of available genome-wide data: (1) genes derived from human GWAS studies: we updated and expanded the GWAS genes catalog from [11]. (2) Genes found in transcriptome studies of rodent models (brain tissue): we obtained the raw expression data from four unique selective-bred rodent models and re-analyzed them to identify strain-specific genes differentially expressed in high- versus low-aggressive lines. (3) Previously published sets of human genes implicated in aggression phenotypes in human single-gene disorders cataloged in online Mendelian inheritance in man (OMIM) [14] and mouse genes implicated in KO studies [15]. This latter category comprises high risk and possibly causal genes because single-gene changes results in (or modifies) the individual’s aggressive behavior. Although each of these studies have intrinsic limitations, the convergence of evidence by cross-referencing and integrating the available data may lead to a more comprehensive and confident understanding of the genetic basis of aggression in humans and mammals.
Material and methods
Aggression gene lists
GWAS gene sets
We updated previous reported GWAS genes for aggression [11] with studies published until August 2016, discarding those that were performed in samples of individuals with other psychiatric disorders (such as drug dependence or BIP). Selected GWAS included four studies for the adult GWAS gene set [32–35] and five for the child gene set [13, 36–39]. Two studies were GWAS meta-analyses [13, 32]. For detailed procedures, see Supplementary Figure 1. Eligible SNPs and retrieved genes are in Supplementary Table 1. As a negative control, we generated a gene list from the GWAS catalog (https://www.ebi.ac.uk/gwas/) by combining signals from 14 phenotypes not related to the nervous system in samples of individuals with European ancestry and sample sizes that are similar to those of the nine GWAS of aggressive behaviors. We used the same procedures to retrieve associated signals and nearby genes (Supplementary Table 2).
Genes from rodent model transcriptomes
Genome-wide transcriptome data were available for four genetic rodent models of aggression: three inbred mouse strains [28] and one rat strain [29] along with their comparable low-aggression strains. Data had been generated using Affymetrix Mouse Genome 430 2.0 or Rat Genome U34 microarrays, and acquired from the Gene Expression Omnibus (dataset series GSE29552) [29] or the author [28]. The mouse strains from the study by Malki et al. were (1) Turku aggressive and Turku non-aggressive mice selected from a colony of Swiss albino mice in Turku (Finland) based on high male–male aggression in a dyadic test against non-aggressive mice [40]; (2) short attack latency and long attack latency mice bred from a wild-type Mus musculus domesticus population in Groningen, Holland by selecting on average attack latency in a resident-intruder test [41]; (3) North Carolina aggressive (NC900) and non-aggressive (NC100) mice selected from out-bred NCR mice in North Carolina (USA) showing increased aggression and reactivity to stimulation [42]. For convenience, we use country of origin to denote the strains: Finland, Holland, and USA, respectively. The rat model data were derived from selectively bred high responder (bHR) and low responder (bLR) Sprague-Dawley rats. These groups showed differences in emotional reactivity and exploratory behavior, aggression, impulsivity, and proclivity to psychostimulant abuse [43, 44]. We used weighted gene co-expression network analysis [45] to identify strain-specific genes in co-expression modules significantly associated with aggression (for details of methods, see Supplementary File 1; gene sets were listed in Supplementary Table 3).
OMIM and KO mice genes
We used previously published sets of human genes implicated in aggression phenotypes in human single-gene disorders cataloged in OMIM (N = 85) [14] and mouse genes implicated in KO studies (N = 89) [15]. Genes in these two sets were included in Supplementary Table 4.
Genetic correlation (LD score) analyses between aggression and psychiatric disorders
We estimated the genetic correlation of aggression with six other psychiatric disorders (ADHD, SCZ, BIP, autism spectrum disorders (ASDs), major depression (MDD) and post-traumatic stress disorders (PTSDs), by LD score (LDSC) regression analysis [46]. We used the largest aggression GWAS meta-analysis of children samples, the EAGLE (Early Genetics and Lifecourse Epidemiology Consortium) study [13] and a recently published GWAS of antisocial behavior by Tielbeek et al. [47], which included 64% adult and 36% child samples. None of the four adult aggression samples reported so far have either sufficient sample sizes or summary statistics available. For ADHD, SCZ, BIP, ASD, MDD, and PTSD, the sources of summary statistics are in Supplementary Table 5.
Gene set overlap analysis and gene ranking
Rodent transcriptome genes were converted to human orthologues using biomaRt [48, 49]. Gene overlap among the sets was evaluated using one-tailed Fisher’s exact tests. We ranked individual genes by their total numbers of occurrences in these lists. We also ranked them using a simple weighted sum method: aggression gene rank = 1 × (total occurrence in human GWAS studies) + 0.5 × (total occurrence in four rodent model transcriptome lists) + 1.5 × (total occurrence in OMIM and KO mice genes). The rationale was to add 50% more weight to the genes in OMIM or in KO mice lists compared with the GWAS genes lists because alterations in the first set of genes are more firmly linked to aggression. We discounted the weight to 0.5 for the rodent transcriptome genes given the limitations of these studies, including the limited phenotypes used in selective breeding, small sample sizes, the limited brain regions and age ranges studied, and the potential for confounding cause and effect in such studies. This weighting scheme also ensured that the maximum possible ranking scores from all four rodent models would be 2, equal to the maximum possible score of the two GWAS lists. The three main categories of studies (human GWAS, rodent transcriptome studies, and the high-risk gene set combining KO mice and OMIM genes) were evaluated for overlap using Fisher’s exact test.
Ingenuity pathway analysis: pathway and network analysis
Individual gene sets were imported to ingenuity pathway analysis (IPA) to assess canonical pathway enrichment. The negative log of Fisher’s exact test p-values are reported. Using a p < 0.05 cutoff, we coded the pathway enrichment as a binary variable for the subsequent analysis, with 1 indicating significant enrichment and 0 no enrichment. We examined the pathway enrichment similarities among the gene sets with classical metric multidimensional scaling (MDS) using the Rogers-Tanmoto correlation for binary data [50] in STATA 14. The configuration for the first four-dimensional Euclidean space was visualized in a 4D plot (a, b, c, and node size) created in R. We explored the activation/inhibition states of the top enriched canonical pathways using IPA’s activation Z-score tool. Because gene expression changes are needed to calculate activation Z-scores and because shared pathway enrichment is needed for the activation comparison, this analysis was only performed on the USA mice and the rat models. Finally, we used IPA’s network generation algorithm to identify the highly interconnected networks of the top 40 ranked genes according to the weighted method described in the previous section. These networks were visualized using IPA’s Path Designer tool. We imported the networks into Cytoscape to measure the number of interactions (degree) of the top-ranked 40 genes with other genes in the network. Logarithms of the degree estimates were compared for rodent versus human aggression genes using quantile regression.
Results
Aggression gene sets
Human GWAS genes
A total of 175 and 281 genes were selected from four adult and five children GWAS gene sets, respectively (Supplementary Figure 1 and Supplementary Table 1). Six genes were present in both sets: ALK, LAMA2, NFKB1, OSMR, RBFOX1, and WDR62 (significant overlap by Fisher’s exact test, p = 0.038, Table 1). The control GWAS gene list comprises 172 genes (Supplementary Table 2); only one gene was shared with either the adult (ARHGEF3) or the child (LY86) datasets (non-significant). LDSC regression found a significant positive correlation between the EAGLE GWAS meta-analysis of aggression in children [13] and ADHD (p = 9.75e−05), and positive correlations between the recently published GWAS meta-analysis of antisocial behavior [47] with ADHD (p = 4.4e−03) and MDD (p = 3.53e−03). No significant genetic correlations were found with any other disorders examined (SCZ, ASD, BIP, and PTSD), or between the aggression GWAS meta-analyses (Supplementary Figure 2).
Table 1.
Gene list overlaps
| Gene overlap | GWAS_Adult | GWAS_Child | KO_Mice | Mice_Finland | Mice_Holland | Mice_USA | OMIM | Rat |
|---|---|---|---|---|---|---|---|---|
| (N/p-values) | (N = 175) | (N = 281) | (N = 89) | (N = 381) | (N = 271) | (N = 397) | (N = 85) | (N = 211) |
| GWAS_Child | 6 | |||||||
| (N = 281) | p = 0.038 | |||||||
| KO_Mice | 1 | 4 | ||||||
| (N = 89) | p = 0.544 | p = 0.037 | ||||||
| Mice_Finland | 1 | 6 | 6 | |||||
| (N = 381) | p = 0.269 | p = 0.662 | p = 0.007 | |||||
| Mice_Holland | 2 | 2 | 1 | 13 | ||||
| (N = 271) | p = 1 | p = 0.597 | p = 1 | p = 0.002 | ||||
| Mice_USA | 3 | 11 | 4 | 11 | 12 | |||
| (N = 397) | p = 1 | p = 0.029 | p = 0.102 | p = 0.193 | p = 0.013 | |||
| OMIM | 1 | 3 | 4 | 3 | 1 | 1 | ||
| (N = 85) | p = 0.528 | p = 0.118 | p = 0.001 | p = 0.221 | p = 1 | p = 1 | ||
| Rat | 1 | 4 | 6 | 5 | 5 | 10 | 3 | |
| (N = 211) | p = 1 | p = 0.546 | p < 0.0001 | p = 0.606 | p = 0.215 | p = 0.010 | p = 0.061 | |
| GWAS_Contrl | 1 | 1 | 2 | 1 | 4 | 2 | 1 | 2 |
| (N = 172) | p = 1 | p = 0.737 | p = 0.179 | p = 0.389 | p = 0.301 | p = 0.779 | p = 0.522 | p = 0.704 |
Rodent transcriptome genes
One gene module was significantly downregulated in bHR rats compared to the bLR rats (ME6, Benjamini-Hochberg (BH)-corrected p < 0.05). For the mouse models, one module for each line was significantly downregulated in high-aggression versus low-aggression lines (Finland: ME12, uncorrected p-value <0.005, BH p-value <0.2; Holland: ME29, BH p < 0.05; USA: ME11, BH p < 0.05). One module was significantly upregulated for the Holland aggressive versus non-aggressive line (ME22, p-value = 8.4e−04, BH p-value = 0.028). Supplementary Figure 3 plots the eigengene expression for all five significant rodent modules.
We combined the two modules for the Holland lines for downstream analyses. This yielded one significantly associated gene set for each single rodent model for a total of four gene sets. The gene sets and eigengene expression for individual genes in each module are shown in Supplementary Table 3. The Holland mice shared 12 genes with the USA lines (p = 0.013) and 13 with the Finland lines (p = 0.002) (Table 1). The Finland and USA lines shared 11 genes (non-significant). The rat model shared five genes with the Finland and Holland lines (non-significant) and 10 with the USA lines (p = 0.01).
Comparison of gene lists and gene ranking
The human GWAS and rodent transcriptome gene sets were cross-referenced with previously published OMIM and KO mice gene sets [15, 51] (Supplementary Table 4). Table 1 summarizes the overlap among all possible pairs and the Fisher’s exact test p-values. Although the number of overlapping genes was small, some reached statistical significance. Notably, the USA mouse shared 11 genes with the child GWAS gene list (p = 0.029), and the KO mice set shared four genes with the child GWAS genes (p = 0.037), four with the human OMIM genes (p = 0.001), six with the Finland mouse (p = 0.007), and six with the rat model (p < 0.001). None of the eight aggression gene lists showed any significant overlap with the GWAS control gene set.
Supplementary Table 4 lists all the 1767 genes from the eight aggression gene sets (adult and children GWAS, transcriptomics in four rodent models, KO mice and OMIM) and ranks them based on their number of occurrences and weighted ranks for aggression (only human orthologs were included from the rodent genes). MAOA was ranked highest with both methods. One hundred and nineteen genes appeared in at least two lists. Forty of them have a weighted ranking score ≥2 (Table 2) and almost all are involved in neuronal functions: synaptic transmission (n = 13, GO: 0007268, p = 5e−09), nervous system development (n = 18, GO: 0007399, p = 7e−08), synapse (n = 11, GO: 0045202, p = 1e−08), neuron projection (n = 9, GO: 0043005, p = 2e−05), and neuroactive ligand–receptor interaction (n = 5, KEGG: 04080, p = 2e−04). Detailed information on individual genes and references to the original studies are shown in Supplementary Table 6.
Table 2.
Top 40 ranked genes
| Gene symbol | Weighted ranking | Gene name | Gene symbol | Weighted ranking | Gene name |
|---|---|---|---|---|---|
| MAOA | 4 | Monoamine oxidase A | ECM1 | 2 | Extracellular matrix protein 1 |
| ERBB4 | 3 | Erb-b2 receptor tyrosine kinase 4 | EEF1A2 | 2 | Eukaryotic translation elongation factor 1 alpha 2 |
| GRIA3 | 3 | Glutamate ionotropic receptor AMPA type subunit 3 | EHMT1 | 2 | Euchromatic histone lysine methyltransferase 1 |
| MECP2 | 3 | Methyl-CpG-binding protein 2 | GAD2 | 2 | Glutamate decarboxylase 2 |
| PRNP | 3 | Prion protein | GDI1 | 2 | GDP dissociation inhibitor 1 |
| AVPR1A | 2.5 | Arginine vasopressin receptor 1A | GRID1 | 2 | Glutamate ionotropic receptor delta type subunit 1 |
| CHMP2B | 2.5 | Charged multivesicular body protein 2B | GRN | 2 | Granulin |
| EN2 | 2.5 | Engrailed homeobox 2 | GSK3A | 2 | Glycogen synthase kinase 3 alpha |
| FGF14 | 2.5 | Fibroblast growth factor 14 | HSF1 | 2 | Heat shock transcription factor 1 |
| HDAC4 | 2.5 | Histone deacetylase 4 | LAMA2 | 2 | Laminin subunit alpha 2 |
| KCNJ18 | 2.5 | Potassium voltage-gated channel subfamily J member 18 | MAPK15 | 2 | Mitogen-activated protein kinase 15 |
| LRRC7 | 2.5 | Leucine rich repeat containing 7 | MME | 2 | Membrane metalloendopeptidase |
| SERPINI1 | 2.5 | Serpin family I member 1 | NFKB1 | 2 | Nuclear factor kappa B subunit 1 |
| ACHE | 2 | Acetylcholinesterase (Cartwright blood group) | NPY1R | 2 | Neuropeptide Y receptor Y1 |
| ALDH5A1 | 2 | Aldehyde dehydrogenase 5 family member A1 | OSMR | 2 | Oncostatin M receptor |
| ALK | 2 | Anaplastic lymphoma receptor tyrosine kinase | PNOC | 2 | Prepronociceptin |
| CACNB3 | 2 | Calcium voltage-gated channel auxiliary subunit beta 3 | RBFOX1 | 2 | RNA-binding protein, fox-1 homolog 1 |
| CADM1 | 2 | Cell adhesion molecule 1 | SPAST | 2 | Spastin |
| CRHR1 | 2 | Corticotropin releasing hormone receptor 1 | SYN1 | 2 | Synapsin I |
| DNAJB5 | 2 | DnaJ heat shock protein family (Hsp40) member B5 | WDR62 | 2 | WD repeat domain 62 |
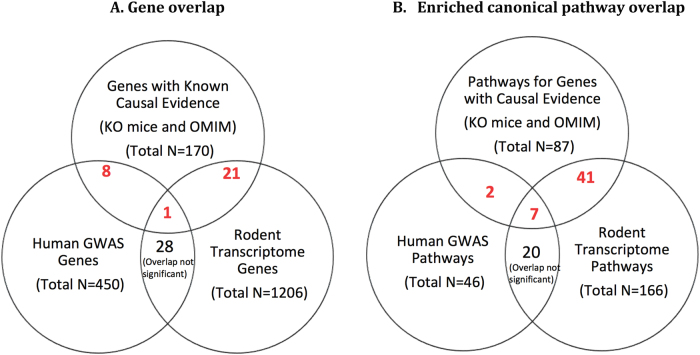
Analysis of overlap among the three main categories human GWAS, rodent transcriptome, and high-risk genes (i.e., KO mice and OMIM genes) revealed one gene, ERBB4, with supporting evidence from all three categories. A total of nine GWAS genes were also high-risk genes (p = 0.007), and 22 rodent transcriptome genes were high-risk genes (p < 0.0001). However, the overlap between GWAS and transcriptome genes (n = 29) was not statistically significant (p = 0.29). The Venn diagram in Fig. 1a shows these overlaps.
Fig. 1.
Venn diagram of gene and pathway overlaps from three categories of genetic evidence. a Number of gene overlaps. One gene, ERBB4, was shared in all three categories. A total of nine GWAS genes were also high-risk genes (p = 0.007), and 22 rodent transcriptome genes were high-risk genes (p < 0.0001). In contrast, the overlap between the human GWAS and the rodent transcriptome genes (n = 29) was not statistically significant (p = 0.29). b Shared canonical pathway enrichments. The total pathway overlap between the human GWAS and high-risk genes (N = 9, p = 0.015) and the overlap between the rodent transcriptome and high-risk genes (N = 48, p = 0.005) were significant. In contrast, the pathway overlap between the human GWAS and rodent transcriptome genes was not significant (N = 27, p = 0.20). Red numbers indicate significant overlaps.
Pathway analysis
We performed canonical pathway enrichment analysis for each aggression gene set. Eleven pathways were significant in at least three aggression gene sets (highlighted in red in Supplementary Table 7) and included axonal guidance signaling, CREB signaling in neurons, ERK/MAPK signaling, G-protein coupled receptor (GPCR) signaling, GABA, and serotonin receptor signaling and reelin signaling. The most shared pathways between any two sets were 11 pathways in common in the OMIM and KO mice gene sets, followed by eight shared pathways between the child GWAS and USA mice and seven between the child GWAS and the rat model. The adult GWAS only shared four pathways with the USA mouse and one with the rat model, although it did share five pathways with the OMIM list. There were no common significantly enriched pathways among all three mouse strains. The Finland and Holland mouse strains had almost no overlap with other gene lists. The child and adult GWAS sets shared only one pathway, phospholipase C signaling. The percentage of enriched pathways shared with any other gene list out of the total enriched canonical pathways for each list were ranked as follows: OMIM 59%, USA mouse 53%, KO mice 48%, Child GWAS and Rat both 44%, adult GWAS 43%, Finland mouse 20% and Holland 11%.
MDS was used to analyze the pathway enrichment similarities shared by the different gene sets. The 4D plot (Supplementary Figure 4 and Supplementary File 2) shows that six sets of aggression genes (two human GWAS, OMIM genes, and all three mouse models) were closely clustered in the first three dimensions, which explained 71.6% of the total variance. The rat model, the KO mice genes, and GWAS control genes were distinctly separable in the first three dimensions. The child GWAS genes mainly loaded on the fourth dimension, which accounted for an additional 12.4% of the variance. The USA mouse model mainly loaded in the fifth dimension, which accounted for an additional 8.6% of the variance (not shown).
Examining the common pathways shared by the three main categories of genetic evidence returned seven canonical pathways (Table 3). Among them, five contained top-ranked genes and the G-protein-coupled receptor signaling pathway was also significantly enriched with the top-ranked genes (p = 0.002). As seen for the analysis of gene overlap, the total pathway overlap between the human GWAS and high-risk genes (N = 9, p = 0.015) and the overlap between the rodent transcriptome and high-risk genes were significant (N = 48, p = 0.005). In contrast, the pathway overlap between the human GWAS and rodent transcriptome genes was not significant (N = 27, p = 0.20). The Venn diagram in Fig. 1b shows this overlap.
Table 3.
Canonical pathways shared by all three categories of genes
| Canonical pathways | Human GWAS genes | Rodent transcriptome genes | Causal genes |
|---|---|---|---|
| Signaling by Rho family GTPases | 2.294 | 1.594 | 1.847 |
| ERK/MAPK signaling | 2.093 | 2.238 | 2.965 |
| G-protein-coupled receptor signaling | 1.999 | 3.971 | 9.286 |
| Axonal guidance signaling | 1.947 | 1.714 | 2.499 |
| Reelin signaling in neurons | 1.939 | 1.759 | 1.432 |
| Molecular mechanisms of cancer | 1.647 | 2.591 | 3.075 |
| Gαs signaling | 1.609 | 2.054 | 5.589 |
The IPA activation Z-score analysis performed on the shared canonical pathways between the USA mice and rat gene sets predicted mostly opposite activities except for one: dopamine DARPP32 feedback in cAMP signaling. This pathway was inhibited in both models (Supplementary Figure 5).
Network analysis of the top genes
The 40 top-ranked genes were highly interconnected in three tightly clustered networks identified by IPA’s network generation algorithm using direct relationships from the Ingenuity® Knowledge Base. These networks were related to nervous system development and function, neurological disease and psychological disorders, and cellular function and maintenance (Fig. 2). The total number of interactions with other genes, i.e, degree, was significantly higher for the human compared with the rodent aggression genes (F(1, 22) = 10.59, p = 0.004). Genes from both human and rodent studies also had a significantly higher degree than the rodent-only genes (F(1, 28) = 4.97, p = 0.034), but this degree is not different from that of the human-only genes (Supplementary Figure 6).
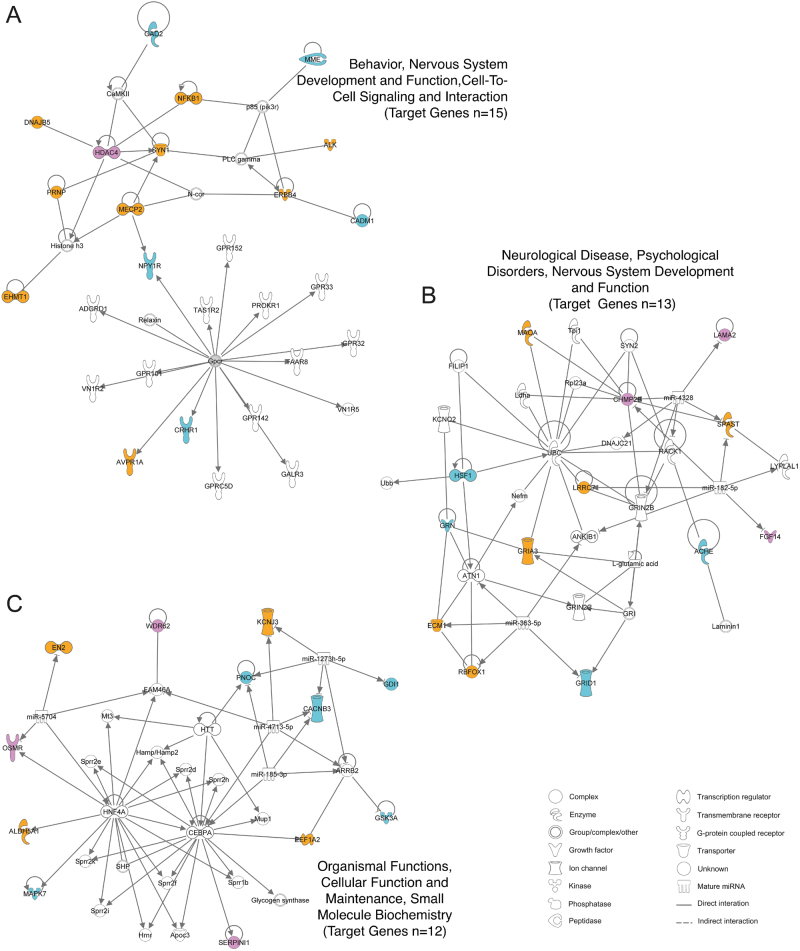
Fig. 2.
IPA network analysis of the 40 top-ranked genes. Network A is involved in behavior, nervous system development and function, and cell-to-cell signaling and interaction (score 35, 15 genes). Network B is involved in neurological disease, psychological disorders, nervous system development and function (score 29, 13 genes). Network C is involved in organismal functions, cellular function and maintenance, small molecule biochemistry (score 26, 12 genes). Genes highlighted in color correspond to the 40 top-ranked genes. In purple, human-only aggression genes (from GWAS and OMIM studies), in blue, rodent-only aggression genes (rodent model transcriptomic genes and KO mice genes), and in orange, genes from both human and rodent studies.
Discussion
Our study integrates all prior human and rodent genetic studies of aggression to overcome their individual limitations and to gain a more robust insight into the genetic etiology of aggression. We found that genes from these different types of studies share some significant overlap at both the gene and biological pathway levels. Their lack of overlap with a control GWAS set confirms that they harbor genetic information relevant to aggression. The two main categories, human GWAS genes and rodent transcriptome genes, however, did not overlap significantly, although they both shared significant overlap with distinct subsets of the high-risk genes (those obtained from human OMIM phenotypes and KO mice). This pattern was also true for the pathway enrichments. This suggests differences in the genetic etiology of human and rodent aggression or the possibility that human GWAS and transcriptomic studies of rodent brains capture different sets of genes involved in the etiology of aggression. Finally, using a weighted ranking method, we provided a hierarchical list of genes associated with aggression.
The adult and child GWAS sets had only six genes in common: ALK, LAMA2, NFKB1, OSMR, RBFOX1, and WDR62. Albeit small, this overlap was statistically significant. All six genes are essential for neurodevelopment. ALK encodes a tyrosine kinase receptor linked to neuroblastoma [52]. LAMA2 encodes an extracellular matrix protein. Its mutation causes denervation atrophy of the muscle [53]. OSMR is a member of the type-I cytokine receptor family and is essential for the development of a subtype of nociceptive neurons in the dorsal root ganglia [54]. NFKB1 is a transcription factor involved in regulating responses of neurons to activation of different signaling pathways in a variety of physiological and pathological conditions [55]. RBFOX1 is a splicing factor implicated in many neurodevelopmental and psychiatric disorders and several evidences have highlighted this gene as a candidate for aggression [56, 57]. WDR62 is a centrosomal and nuclear protein linked to autosomal recessive microcephaly [58, 59].
Most genes in the adult and child GWAS sets did not overlap. The two sets only shared one canonical pathway. Furthermore, the EAGLE GWAS meta-analysis of children’s aggression [13] and the Tielbeek’s GWAS of antisocial behavior (64% adult) [47] do not show genetic correlation. The sample size of children in Tielbeek’s GWAS was only half of the EAGLE’s, likely too small for LDSC regression and explaining their lack of correlation despite the presence of children samples in both datasets. More importantly, the results highlight the possible genetic differences, although it may be due to phenotype differences between the two samples. Indeed, aggressive behaviors often manifest in different forms and are triggered by different risk factors across the lifespan [60]. Some aggressive behaviors in childhood predict aggression in later life; others are temporary and disappear at later ages, such as temper tantrums in toddlers [61] and adolescence-limited antisocial behavior [62]. Adult onset aggression is often linked to physical or emotional trauma, substance use, medical illnesses, or brain injuries [60]. Our LDSC regression found significant correlations between the EAGLE’s child sample and ADHD, and between Tielbeek’s sample (64% adults) with both ADHD and MDD. The results support pleiotropic effects of shared common DNA variants on the comorbidity of aggression with ADHD in children, or with MDD in adults. The lack of genetic correlations of either dataset with SCZ, BIP, autism, or PTSD suggests that for these disorders, aggression may arise from different causal factors. Future studies are needed to fully address the genetic bases of the comorbidity between aggression and psychiatric disorders.
Among the rodent models, few biological pathways were shared, although there was some significant gene overlap. Differences in selective breeding may have fixed different genes into these models. Considering that the Finland and Holland lines were selectively bred based on a single behavioral criterion, either increased aggression toward non-aggressive mice [40] or decreased attack latency toward intruder mice [41], it is not surprising to see that they had the lowest numbers of enriched pathways and overlap with other lists. Our results suggest that their utility in modeling human aggression may be limited because the underlying genetic risk factors may be different from one another and more importantly from the genetic predisposition to aggression in humans. This notion was further supported by our network analysis of the 40 top-ranked genes, which showed that genes with evidence only from rodent studies have a fewer number of interactions in the network than those genes derived from human studies, suggesting more restricted functional impact of rodent genes on the network.
In contrast, the USA mice and the rat lines showed many overlapping pathways with the human GWAS genes, most notably with the child set. The USA mice and the rat model were bred for varying phenotypes: increased reactivity toward stimulation [42], novelty exploration, impulsivity, and vulnerability to psychostimulant abuse [29, 44]. Considering that human aggression is often accompanied by these traits, the USA mouse (NC900/NC100) and rat (bHR/bLR) lines may be better suited to model human aggression, particularly in the context of psychiatric comorbidities. However, their shared biological pathways often showed opposite inhibition/activation activities and, although statistically significant, the two lines shared <5% of genes (Table 1). Thus, it seems likely that different mechanisms, having some shared components, regulate aggression in these two models. The only consistent directional change of these shared pathways between the two rodent models, the downregulation of the dopamine-DARPP32 feedback in cAMP signaling, was also found altered in the ventral striatum and the frontal cortex of an operant mouse model of frustration showing aggressive behavior [63].
The most notable overlap observed in our analyses was for the 11 pathways shared between the human OMIM and the KO mice genes. Because the OMIM phenotypes are typically multidimensional phenotypes including medical, psychiatric, and aggressive features, it has not heretofore been clear whether the aggression observed in affected individuals was a direct genetic effect or if it was mediated via another phenotype. The substantial overlap with the KO mouse gene set provides strong evidence that the aggression in these OMIM disorders has a genetic etiology.
Among the top enriched pathways, several were previously well-known pathways for aggression: the dopamine, serotonin, glutamate, and GABA signaling pathways (Supplementary Table 7). Some of the pathways we found were not previously linked to aggression directly. However, there are several reasons to view them as functionally associated with aggression. For example, the GPCR signaling pathway, which was significantly enriched in our data (Table 3), mediates much receptor signaling including serotonin, dopamine, metabolic glutamate receptors, oxytocin and vasopressin receptors. ERK/MAPK and Rho-GTPase signaling form intracellular signaling cascades that orchestrate cellular responses of GPCR signaling. Axonal guidance and reelin signaling are important pathways for nervous system development and have been implicated in neuropsychiatric disorders such as bipolar, SCZ, and ADHD [64–68], which are often associated with aggression. Indeed, several recent reviews in human and animals have consistently identified these pathways [11, 15, 51]. Novel pathways that have never been linked to aggression offer us new perspectives on the pathophysiology of aggression. One interesting example is cancer signaling. Although it has never been implicated in aggression, it is not uncommon for cancer patients to display changed personalities and even violent behaviors [69]. The close relationship of cancer signaling with immune system offers a plausible mechanism linking cancer with many neuropsychiatric conditions including aggression.
Finally, one notable finding is that our ranked gene list highlights 40 top genes (Table 2 and Supplementary Table 6), all of which are involved in neurotransmission, axon guidance, synaptic plasticity, learning and memory, neuronal development, or hormone signaling. Twenty-three of the top genes had reports of KO mice studies, strongly supporting their role in aggression. One particular gene of our interest is RBFOX1, a splicing factor important for neuronal development. Interestingly, the protein encoded by RBFOX1 regulates the expression of 15 of the top 40 ranked genes (the probability of this event is p = 3.4e−05) [70]. Convergent data from GWAS, neuroimaging genetics, epigenetics, gene expression, and animal models supports RBFOX1 as a strong candidate for aggression [57]. Furthermore, all 40 genes are highly connected in three functional networks (Fig. 2). Human disease genes tend to interact with each other with higher network connectivity than non-disease genes [71]. Many studies of complex neuropsychiatric disorders have also concluded that disease-causing variants are often clustered in protein-interaction networks with a high degree of connectivity among themselves and that these clustered networks are often enriched with functional pathways relevant to brain functions [72–80]. We observed both characteristics for our top-ranked genes, which strongly supports a multifactorial genetic landscape for aggression and the roles of these top genes in aggression. The clusters formed by the top-ranked genes are not only crucial keys for deciphering molecular mechanisms underlying the pathophysiology of aggression; they may also harbor useful therapeutic targets.
We noted that non-genetic models, such as stress-induced aggression [27, 81] and other organisms [30, 31, 82], were not included in this study. However, some correspondences with our findings are worth mentioning (details in Supplementary File 1). These consistencies support the utility of a cross-species approach like ours for identifying genetic mechanisms that are evolutionarily conserved and that may underlie gene-by-environment interactions.
Our approach inherits the limitations of the original studies. GWAS were underpowered and the resulting gene sets may include many false positives. KO mice studies are biased by authors’ choices. For example, genes reported to cause aggression in OMIM disorders may be more likely selected for gene KO in animal models. Although these KO models validate the role of these genes in aggression, it limits our ability to understand the true degree of overlap. The rodent genetic models had been defined by simple behavioral criteria, which may not be generalizable to other species. Improving aggression studies, for example by increasing GWAS sample sizes or by building a repository of behavioral phenotypes for gene KOs in mice, could certainly improve accuracy and decrease the noise in integrative studies like ours. Including other species and model organisms may also provide additional insights; however, difficulty remains regarding gene orthology and generalization of behaviors across distant species.
In summary, we integrated genomic and transcriptomic studies from different species and provided valuable insights into the complex genetic signatures that underlie aggression in both humans and rodent models. Our ranked lists of genes and pathways provide guidance for functional studies in the future.
Electronic supplementary material
Acknowledgements
S.V.F. is supported by the K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway, the European Union’s Seventh Framework Programme [FP7/2007–2013] for research, technological development and demonstration under grant agreement number 602805, the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 667302, and NIMH grants 5R01MH101519 and U01 MH109536-01.
Funding
Financial support was received by B.C. from the Spanish “Ministerio de Economía y Competitividad” (SAF2015-68341-R) and from “Generalitat de Catalunya-AGAUR” (2017-SGR-738). The research leading to these results has also received funding from the European Union Seventh Framework Programme [FP7/2007–2013] under grant agreement no. 602805, from the European Union H2020 Programme [H2020/2014-2020] under grant agreement no. 667302 and from the European College for Neuropsychopharmacology (ECNP) by a grant for the ECNP Network “ADHD across the lifespan.” N.F.-C. were supported by a contract from the “Centro de Investigación Biomédica en Red de Enfermedades Raras” (CIBERER). Y.Z.-J. is supported by the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 602805, and the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 667302.
Compliance with ethical standards
Conflict of interest
In the past year, S.V.F. received income, travel expenses, and/or research support from and/or has been on an Advisory Board for Pfizer, Ironshore, Shire, Akili Interactive Labs, CogCubed, Alcobra, VAYA Pharma, Neurovance, Impax, NeuroLifeSciences, and research support from the National Institutes of Health (NIH). With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. In previous years, he received consulting fees or was on Advisory Boards or participated in continuing medical education programs sponsored by: Shire, Alcobra, Otsuka, McNeil, Janssen, Novartis, Pfizer, and Eli Lilly. S.V.F. receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts, and Elsevier: ADHD: Non-Pharmacologic Treatments. During the past year, K.M. received income from and has been an employee and stockholder of Eli Lilly and UCB Celltech. The remaining authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41380-018-0068-7) contains supplementary material, which is available to authorized users.
References
- 1.Retz W, Rosler M. The relation of ADHD and violent aggression: what can we learn from epidemiological and genetic studies? Int J Law Psychiatry. 2009;32:235–43. doi: 10.1016/j.ijlp.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 2.McKay KE, Halperin JM. ADHD, aggression, and antisocial behavior across the lifespan. Interactions with neurochemical and cognitive function. Ann N Y Acad Sci. 2001;931:84–96. doi: 10.1111/j.1749-6632.2001.tb05774.x. [DOI] [PubMed] [Google Scholar]
- 3.Manchia M, Fanos V. Targeting aggression in severe mental illness: the predictive role of genetic, epigenetic, and metabolomic markers. Prog Neuropsychopharmacol Biol Psychiatry. 2017;77:32–41. doi: 10.1016/j.pnpbp.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Bulgari V, Iozzino L, Ferrari C, Picchioni M, Candini V, De Francesco A, et al. Clinical and neuropsychological features of violence in schizophrenia: a prospective cohort study. Schizophr Res. 2017;181:124–30. doi: 10.1016/j.schres.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Volavka J. Violence in schizophrenia and bipolar disorder. Psychiatr Danub. 2013;25:24–33. [PubMed] [Google Scholar]
- 6.Tuvblad C, Baker LA. Human aggression across the lifespan: genetic propensities and environmental moderators. Adv Genet. 2011;75:171–214. doi: 10.1016/B978-0-12-380858-5.00007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Liu C, Chen C, Moyzis R, Chen W, Dong Q. Genetic variations in the serotoninergic system and environmental factors contribute to aggressive behavior in Chinese adolescents. Physiol Behav. 2015;138:62–8. doi: 10.1016/j.physbeh.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Ohlsson H, Morris NA, Sundquist J, Sundquist K. A Swedish population-based study of the mechanisms of parent-offspring transmission of criminal behavior. Psychol Med. 2015;45:1093–102. doi: 10.1017/S0033291714002268. [DOI] [PubMed] [Google Scholar]
- 9.Laucht M, Brandeis D, Zohsel K. Gene-environment interactions in the etiology of human violence. Curr Top Behav Neurosci. 2014;17:267–95. doi: 10.1007/7854_2013_260. [DOI] [PubMed] [Google Scholar]
- 10.Waltes R, Chiocchetti AG, Freitag CM. The neurobiological basis of human aggression: a review on genetic and epigenetic mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2016;171:650–75. doi: 10.1002/ajmg.b.32388. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Castillo N, Cormand B. Aggressive behavior in humans: genes and pathways identified through association studies. Am J Med Genet B Neuropsychiatr Genet. 2016;171:676–96. doi: 10.1002/ajmg.b.32419. [DOI] [PubMed] [Google Scholar]
- 12.Vassos E, Collier DA, Fazel S. Systematic meta-analyses and field synopsis of genetic association studies of violence and aggression. Mol Psychiatry. 2014;19:471–7. doi: 10.1038/mp.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pappa I, St Pourcain B, Benke K, Cavadino A, Hakulinen C, Nivard MG, et al. A genome-wide approach to children’s aggressive behavior: the EAGLE consortium. Am J Med Genet B Neuropsychiatr Genet. 2016;5:562–72. [DOI] [PubMed]
- 14.Zhang-James Y, Faraone SV. Genetic architecture for human aggression: a study of gene-phenotype relationship in OMIM. Am J Med Genet B Neuropsychiatr Genet. 2016;5:641–9. [DOI] [PubMed]
- 15.Veroude K, Zhang-James Y, Fernandez-Castillo N, Bakker MJ, Cormand B, Faraone SV. Genetics of aggressive behavior: an overview. Am J Med Genet B Neuropsychiatr Genet. 2016;1:3–43. [DOI] [PubMed]
- 16.Nelson RJ, Chiavegatto S. Aggression in knockout mice. ILAR J. 2000;41:153–62. doi: 10.1093/ilar.41.3.153. [DOI] [PubMed] [Google Scholar]
- 17.de Boer SF, Olivier B, Veening J, Koolhaas JM. The neurobiology of offensive aggression: revealing a modular view. Physiol Behav. 2015;146:111–27. doi: 10.1016/j.physbeh.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 18.Miczek KA, de Almeida RM, Kravitz EA, Rissman EF, de Boer SF, Raine A. Neurobiology of escalated aggression and violence. J Neurosci. 2007;27:11803–6. doi: 10.1523/JNEUROSCI.3500-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomot M, Gendrot C, Verloes A, Raynaud M, David A, Yntema HG, et al. MECP2 gene mutations in non-syndromic X-linked mental retardation: phenotype-genotype correlation. Am J Med Genet A. 2003;123A:129–39. doi: 10.1002/ajmg.a.20247. [DOI] [PubMed] [Google Scholar]
- 20.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–58. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nitrini R, Rosemberg S, Passos-Bueno MR, da Silva LS, Iughetti P, Papadopoulos M, et al. Familial spongiform encephalopathy associated with a novel prion protein gene mutation. Ann Neurol. 1997;42:138–46. doi: 10.1002/ana.410420203. [DOI] [PubMed] [Google Scholar]
- 22.Budefeld T, Majer A, Jerin A, Majdic G. Deletion of the prion gene Prnp affects offensive aggression in mice. Behav Brain Res. 2014;266:216–21. doi: 10.1016/j.bbr.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz M, Zafar S, Silva CJ, Zerr I. Behavioral abnormalities in prion protein knockout mice and the potential relevance of PrP(C) for the cytoskeleton. Prion. 2014;8:381–6. doi: 10.4161/19336896.2014.983746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mick E, Byrne D, Fried R, Monuteaux M, Faraone SV, Biederman J. Predictors of ADHD persistence in girls at 5-year follow-up. J Atten Disord. 2011;15:183–92. doi: 10.1177/1087054710362217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vessey JP, Karra D. More than just synaptic building blocks: scaffolding proteins of the post-synaptic density regulate dendritic patterning. J Neurochem. 2007;102:324–32. doi: 10.1111/j.1471-4159.2007.04662.x. [DOI] [PubMed] [Google Scholar]
- 26.Carlisle HJ, Luong TN, Medina-Marino A, Schenker L, Khorosheva E, Indersmitten T, et al. Deletion of densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J Neurosci. 2011;31:16194–207. doi: 10.1523/JNEUROSCI.5877-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malki K, Tosto MG, Pain O, Sluyter F, Mineur YS, Crusio WE, et al. Comparative mRNA analysis of behavioral and genetic mouse models of aggression. Am J Med Genet B Neuropsychiatr Genet. 2016;171B:427–36. doi: 10.1002/ajmg.b.32424. [DOI] [PubMed] [Google Scholar]
- 28.Malki K, Pain O, Du Rietz E, Tosto MG, Paya-Cano J, Sandnabba KN, et al. Genes and gene networks implicated in aggression related behaviour. Neurogenetics. 2014;15:255–66. doi: 10.1007/s10048-014-0417-x. [DOI] [PubMed] [Google Scholar]
- 29.Clinton SM, Stead JD, Miller S, Watson SJ, Akil H. Developmental underpinnings of differences in rodent novelty-seeking and emotional reactivity. Eur J Neurosci. 2011;34:994–1005. doi: 10.1111/j.1460-9568.2011.07811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malki K, Du Rietz E, Crusio WE, Pain O, Paya-Cano J, Karadaghi RL, et al. Transcriptome analysis of genes and gene networks involved in aggressive behavior in mouse and zebrafish. Am J Med Genet B Neuropsychiatr Genet. 2016;171:827–38. doi: 10.1002/ajmg.b.32451. [DOI] [PubMed] [Google Scholar]
- 31.Kudo A, Shigenobu S, Kadota K, Nozawa M, Shibata TF, Ishikawa Y, et al. Comparative analysis of the brain transcriptome in a hyper-aggressive fruit fly, Drosophila prolongata. Insect Biochem Mol Biol. 2017;82:11–20. doi: 10.1016/j.ibmb.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Brevik EJ, van Donkelaar MM, Weber H, Sanchez-Mora C, Jacob C, Rivero O, et al. Genome-wide analyses of aggressiveness in attention-deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2016;171:733–47. doi: 10.1002/ajmg.b.32434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merjonen P, Keltikangas-Jarvinen L, Jokela M, Seppala I, Lyytikainen LP, Pulkki-Raback L, et al. Hostility in adolescents and adults: a genome-wide association study of the Young Finns. Transl Psychiatry. 2011;1:e11. doi: 10.1038/tp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mick E, McGough J, Deutsch CK, Frazier JA, Kennedy D, Goldberg RJ. Genome-wide association study of proneness to anger. PLoS ONE. 2014;9:e87257. doi: 10.1371/journal.pone.0087257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tielbeek JJ, Medland SE, Benyamin B, Byrne EM, Heath AC, Madden PA, et al. Unraveling the genetic etiology of adult antisocial behavior: a genome-wide association study. PLoS ONE. 2012;7:e45086. doi: 10.1371/journal.pone.0045086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aebi M, van Donkelaar MM, Poelmans G, Buitelaar JK, Sonuga-Barke EJ, Stringaris A, et al. Gene-set and multivariate genome-wide association analysis of oppositional defiant behavior subtypes in attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2016;171:573–88. doi: 10.1002/ajmg.b.32346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anney RJ, Lasky-Su J, O’Dushlaine C, Kenny E, Neale BM, Mulligan A, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1369–78. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- 38.Mick E, McGough J, Loo S, Doyle AE, Wozniak J, Wilens TE, et al. Genome-wide association study of the child behavior checklist dysregulation profile. J Am Acad Child Adolesc Psychiatry. 2011;50:807–17.e8. doi: 10.1016/j.jaac.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viding E, Hanscombe KB, Curtis CJ, Davis OS, Meaburn EL, Plomin R. In search of genes associated with risk for psychopathic tendencies in children: a two-stage genome-wide association study of pooled DNA. J Child Psychol Psychiatry. 2010;51:780–8. doi: 10.1111/j.1469-7610.2010.02236.x. [DOI] [PubMed] [Google Scholar]
- 40.Lagerspetz K. Genetic and social causes of aggressive behaviour in mice. Scand J Psychol. 1961;2:167–73. doi: 10.1111/j.1467-9450.1961.tb01237.x. [DOI] [Google Scholar]
- 41.van Oortmerssen GA, Bakker TC. Artificial selection for short and long attack latencies in wild Mus musculus domesticus. Behav Genet. 1981;11:115–26. doi: 10.1007/BF01065622. [DOI] [PubMed] [Google Scholar]
- 42.Sandnabba NK. Differences between two strains of mice, selectively bred for high and low aggressiveness, in the capacity of male odors to affect aggressive behavior. Aggress Behav. 1986;12:103–10. doi: 10.1002/1098-2337(1986)12:2<103::AID-AB2480120204>3.0.CO;2-H. [DOI] [Google Scholar]
- 43.Stead JD, Clinton S, Neal C, Schneider J, Jama A, Miller S, et al. Selective breeding for divergence in novelty-seeking traits: heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet. 2006;36:697–712. doi: 10.1007/s10519-006-9058-7. [DOI] [PubMed] [Google Scholar]
- 44.Flagel SB, Robinson TE, Clark JJ, Clinton SM, Watson SJ, Seeman P, et al. An animal model of genetic vulnerability to behavioral disinhibition and responsiveness to reward-related cues: implications for addiction. Neuropsychopharmacology. 2010;35:388–400. doi: 10.1038/npp.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tielbeek JJ, Johansson A, Polderman TJC, Rautiainen MR, Jansen P, Taylor M, et al. Genome-wide association studies of a broad spectrum of antisocial behavior. JAMA Psychiatry. 2017;12:1242–50. [DOI] [PMC free article] [PubMed]
- 48.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–40. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 49.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Srihari SN. Properties of binary vector dissimilarity measures. InProc. JCIS Int’l Conf. computer vision, pattern recognition, and image processing 2003; Vol. 1.
- 51.Zhang-James Y, Faraone SV. Genetic architecture for human aggression - a study of gene-phenotype relationship in OMIM. World Congress of Psychiatric Genetics - WCPG 2015: Toronto; 2015. [DOI] [PubMed]
- 52.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall TE, Bryson-Richardson RJ, Berger S, Jacoby AS, Cole NJ, Hollway GE, et al. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc Natl Acad Sci USA. 2007;104:7092–7. doi: 10.1073/pnas.0700942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morikawa Y, Tamura S, Minehata K, Donovan PJ, Miyajima A, Senba E. Essential function of oncostatin m in nociceptive neurons of dorsal root ganglia. J Neurosci. 2004;24:1941–7. doi: 10.1523/JNEUROSCI.4975-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattson MP. NF-kappaB in the survival and plasticity of neurons. Neurochem Res. 2005;30:883–93. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- 56.Bill BR, Lowe JK, Dybuncio CT, Fogel BL. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int Rev Neurobiol. 2013;113:251–67. doi: 10.1016/B978-0-12-418700-9.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez-Castillo N, Gan G, van Donkelaar MMJ, Vaht M, Weber H, Retz W, et al. RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior. Eur Neuropsychopharmacol. 2017;6:532–40. [DOI] [PMC free article] [PubMed]
- 58.Bhat V, Girimaji SC, Mohan G, Arvinda HR, Singhmar P, Duvvari MR, et al. Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clin Genet. 2011;80:532–40. doi: 10.1111/j.1399-0004.2011.01686.x. [DOI] [PubMed] [Google Scholar]
- 59.Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–4. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Lewis G, Evans L. Understanding aggressive behaviour across the lifespan. J Psychiatr Ment Health Nurs. 2013;20:156–68. doi: 10.1111/j.1365-2850.2012.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keenan K, Wakschlag LS. More than the terrible twos: the nature and severity of behavior problems in clinic-referred preschool children. J Abnorm Child Psychol. 2000;28:33–46. doi: 10.1023/A:1005118000977. [DOI] [PubMed] [Google Scholar]
- 62.Moffitt TE, Caspi A. Childhood predictors differentiate life-course persistent and adolescence-limited antisocial pathways among males and females. Dev Psychopathol. 2001;13:355–75. doi: 10.1017/S0954579401002097. [DOI] [PubMed] [Google Scholar]
- 63.Martin-Garcia E, Fernandez-Castillo N, Burokas A, Gutierrez-Cuesta J, Sanchez-Mora C, Casas M, et al. Frustrated expected reward induces differential transcriptional changes in the mouse brain. Addict Biol. 2015;20:22–37. doi: 10.1111/adb.12188. [DOI] [PubMed] [Google Scholar]
- 64.Mooney MA, McWeeney SK, Faraone SV, Hinney A, Hebebrand J, Consortium I, et al. Pathway analysis in attention deficit hyperactivity disorder: an ensemble approach. Am J Med Genet B Neuropsychiatr Genet. 2016;171:815–26. doi: 10.1002/ajmg.b.32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen N, Bao Y, Xue Y, Sun Y, Hu D, Meng S, et al. Meta-analyses of RELN variants in neuropsychiatric disorders. Behav Brain Res. 2017;332:110–9. doi: 10.1016/j.bbr.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 66.Li W, Guo X, Xiao S. Evaluating the relationship between reelin gene variants (rs7341475 and rs262355) and schizophrenia: a meta-analysis. Neurosci Lett. 2015;609:42–7. doi: 10.1016/j.neulet.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 67.Chen SY, Huang PH, Cheng HJ. Disrupted-in-schizophrenia 1-mediated axon guidance involves TRIO-RAC-PAK small GTPase pathway signaling. Proc Natl Acad Sci USA. 2011;108:5861–6. doi: 10.1073/pnas.1018128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goes FS, Willour VL, Zandi PP, Belmonte PL, MacKinnon DF, Mondimore FM, et al. Sex-specific association of the reelin gene with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:549–53. doi: 10.1002/ajmg.b.31018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grube M. Violent behavior in cancer patients--a rarely addressed phenomenon in oncological treatment. J Interpers Violence. 2012;27:2163–82. doi: 10.1177/0886260511431434. [DOI] [PubMed] [Google Scholar]
- 70.Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, et al. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–28. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ben-David E, Shifman S. Networks of neuronal genes affected by common and rare variants in autism spectrum disorders. PLoS Genet. 2012;8:e1002556. doi: 10.1371/journal.pgen.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakai Y, Shaw CA, Dawson BC, Dugas DV, Al-Mohtaseb Z, Hill DE, et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med. 2011;3:86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanz TA, Guilmette E, Gosink MM, Fischer JE, Fitzgerald LW, Stephenson DT, et al. Transcriptomic analysis of genetically defined autism candidate genes reveals common mechanisms of action. Mol Autism. 2013;4:45. doi: 10.1186/2040-2392-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry. 2011;168:365–77. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- 80.Yang L, Neale BM, Liu L, Lee SH, Wray NR, Ji N, et al. Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. Am J Med Genet B Neuropsychiatr Genet. 2013;162:419–30. doi: 10.1002/ajmg.b.32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muhie S, Gautam A, Meyerhoff J, Chakraborty N, Hammamieh R, Jett M. Brain transcriptome profiles in mouse model simulating features of post-traumatic stress disorder. Mol Brain. 2015;8:14. doi: 10.1186/s13041-015-0104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zapata I, Serpell JA, Alvarez CE. Genetic mapping of canine fear and aggression. BMC Genomics. 2016;17:572. doi: 10.1186/s12864-016-2936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.