Structure of linkage disequilibrium and phenotypic associations in the maize genome - PubMed
- ️Mon Jan 01 2001
Structure of linkage disequilibrium and phenotypic associations in the maize genome
D L Remington et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Association studies based on linkage disequilibrium (LD) can provide high resolution for identifying genes that may contribute to phenotypic variation. We report patterns of local and genome-wide LD in 102 maize inbred lines representing much of the worldwide genetic diversity used in maize breeding, and address its implications for association studies in maize. In a survey of six genes, we found that intragenic LD generally declined rapidly with distance (r(2) < 0.1 within 1500 bp), but rates of decline were highly variable among genes. This rapid decline probably reflects large effective population sizes in maize during its evolution and high levels of recombination within genes. A set of 47 simple sequence repeat (SSR) loci showed stronger evidence of genome-wide LD than did single-nucleotide polymorphisms (SNPs) in candidate genes. LD was greatly reduced but not eliminated by grouping lines into three empirically determined subpopulations. SSR data also supplied evidence that divergent artificial selection on flowering time may have played a role in generating population structure. Provided the effects of population structure are effectively controlled, this research suggests that association studies show great promise for identifying the genetic basis of important traits in maize with very high resolution.
Figures
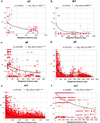
Plots of squared correlations of allele frequencies (r2) against weighted distance between polymorphic sites in six candidate genes: (a) id1, (b) tb1, (c) d8, (d) d3, (e) sh1, and (f) su1. Curves show nonlinear regression of r2 on weighted distance, by using a recombination-drift model for su1 and a mutation-recombination drift model for all other loci. Regression coefficients (b1) and the corrected percentage of variance explained by the models (SSM/SSC) are shown above each plot.
Similar articles
-
Linkage disequilibrium and haplotype block patterns in popcorn populations.
Andrade ACB, Viana JMS, Pereira HD, Pinto VB, Fonseca E Silva F. Andrade ACB, et al. PLoS One. 2019 Sep 25;14(9):e0219417. doi: 10.1371/journal.pone.0219417. eCollection 2019. PLoS One. 2019. PMID: 31553737 Free PMC article.
-
Comprehensive genotyping of the USA national maize inbred seed bank.
Romay MC, Millard MJ, Glaubitz JC, Peiffer JA, Swarts KL, Casstevens TM, Elshire RJ, Acharya CB, Mitchell SE, Flint-Garcia SA, McMullen MD, Holland JB, Buckler ES, Gardner CA. Romay MC, et al. Genome Biol. 2013 Jun 11;14(6):R55. doi: 10.1186/gb-2013-14-6-r55. Genome Biol. 2013. PMID: 23759205 Free PMC article.
-
Thirunavukkarasu N, Hossain F, Shiriga K, Mittal S, Arora K, Rathore A, Mohan S, Shah T, Sharma R, Namratha PM, Mithra AS, Mohapatra T, Gupta HS. Thirunavukkarasu N, et al. BMC Genomics. 2013 Dec 13;14:877. doi: 10.1186/1471-2164-14-877. BMC Genomics. 2013. PMID: 24330649 Free PMC article.
-
Genetic association mapping and genome organization of maize.
Yu J, Buckler ES. Yu J, et al. Curr Opin Biotechnol. 2006 Apr;17(2):155-60. doi: 10.1016/j.copbio.2006.02.003. Epub 2006 Feb 28. Curr Opin Biotechnol. 2006. PMID: 16504497 Review.
-
Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize.
Sahito JH, Zhang H, Gishkori ZGN, Ma C, Wang Z, Ding D, Zhang X, Tang J. Sahito JH, et al. Int J Mol Sci. 2024 Feb 5;25(3):1918. doi: 10.3390/ijms25031918. Int J Mol Sci. 2024. PMID: 38339196 Free PMC article. Review.
Cited by
-
Abundant microsatellite diversity and oil content in wild Arachis species.
Huang L, Jiang H, Ren X, Chen Y, Xiao Y, Zhao X, Tang M, Huang J, Upadhyaya HD, Liao B. Huang L, et al. PLoS One. 2012;7(11):e50002. doi: 10.1371/journal.pone.0050002. Epub 2012 Nov 20. PLoS One. 2012. PMID: 23185514 Free PMC article.
-
Identification of Genetic Loci Associated with Quality Traits in Almond via Association Mapping.
Font i Forcada C, Oraguzie N, Reyes-Chin-Wo S, Espiau MT, Socias i Company R, Fernández i Martí A. Font i Forcada C, et al. PLoS One. 2015 Jun 25;10(6):e0127656. doi: 10.1371/journal.pone.0127656. eCollection 2015. PLoS One. 2015. PMID: 26111146 Free PMC article.
-
Bhat JA, Adeboye KA, Ganie SA, Barmukh R, Hu D, Varshney RK, Yu D. Bhat JA, et al. Front Genet. 2022 Aug 17;13:953833. doi: 10.3389/fgene.2022.953833. eCollection 2022. Front Genet. 2022. PMID: 36419833 Free PMC article.
-
SNP genotyping in melons: genetic variation, population structure, and linkage disequilibrium.
Esteras C, Formisano G, Roig C, Díaz A, Blanca J, Garcia-Mas J, Gómez-Guillamón ML, López-Sesé AI, Lázaro A, Monforte AJ, Picó B. Esteras C, et al. Theor Appl Genet. 2013 May;126(5):1285-303. doi: 10.1007/s00122-013-2053-5. Epub 2013 Feb 5. Theor Appl Genet. 2013. PMID: 23381808
-
Fricano A, Bakaher N, Del Corvo M, Piffanelli P, Donini P, Stella A, Ivanov NV, Pozzi C. Fricano A, et al. BMC Genet. 2012 Mar 21;13:18. doi: 10.1186/1471-2156-13-18. BMC Genet. 2012. PMID: 22435796 Free PMC article.
References
-
- Stuber C W, Polacco M, Senior M L. Crop Sci. 1999;39:1571–1583.
-
- Lai C, Lyman R F, Long A D, Langley C H, Mackay T F C. Science. 1994;266:1697–1702. - PubMed
-
- Laitinen T, Kauppi P, Ignatius J, Ruotsalainen T, Daly M J, Kaariainen H, Kruglyak L, Laitinen H, de la Chapelle A, Lander E S, Laitinen L A, Kere J. Hum Mol Genet. 1997;6:2069–2076. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials