Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics - PubMed
- ️Tue Jan 01 2002
Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics
Sylwia Wasiak et al. J Cell Biol. 2002.
Abstract
Despite numerous advances in the identification of the molecular machinery for clathrin-mediated budding at the plasma membrane, the mechanistic details of this process remain incomplete. Moreover, relatively little is known regarding the regulation of clathrin-mediated budding at other membrane systems. To address these issues, we have utilized the powerful new approach of subcellular proteomics to identify novel proteins present on highly enriched clathrin-coated vesicles (CCVs). Among the ten novel proteins identified is the rat homologue of a predicted gene product from human, mouse, and Drosophila genomics projects, which we named enthoprotin. Enthoprotin is highly enriched on CCVs isolated from rat brain and liver extracts. In cells, enthoprotin demonstrates a punctate staining pattern that is concentrated in a perinuclear compartment where it colocalizes with clathrin and the clathrin adaptor protein (AP)1. Enthoprotin interacts with the clathrin adaptors AP1 and with Golgi-localized, gamma-ear-containing, Arf-binding protein 2. Through its COOH-terminal domain, enthoprotin binds to the terminal domain of the clathrin heavy chain and stimulates clathrin assembly. These data suggest a role for enthoprotin in clathrin-mediated budding on internal membranes. Our study reveals the utility of proteomics in the identification of novel vesicle trafficking proteins.
Figures

Identification of enthoprotin as a novel CCV-associated protein. (A) Proteins of CCVs from rat brain were separated on SDS-PAGE and stained with Coomassie blue. The dashes and brackets indicate the migratory positions of major CCV proteins including the clathrin heavy chain (CHC), the large subunits of the AP1 and AP2 adaptor complexes (α/β/γ-adaptins), the μ-adaptins, σ2-adaptin, and the clathrin light chains (CLCs). (B) Homogenates and microsomes from nontransfected (NT) COS-7 cells and homogenates from cells transfected with FLAG-tagged full-length enthoprotin (T) were analyzed by Western blot with antibodies against enthoprotin (3194) or the FLAG epitope tag as indicated. (C) CCVs were prepared from rat brain and liver and the distribution of endogenous enthoprotin in the subcellular fractions was determined by Western blot and compared with that of endogenous clathrin. H, homogenate; P, pellet; S, supernatant; SGp, sucrose gradient pellet; SGs, sucrose gradient supernatant. (D) A crude preparation of CCVs was generated from HEK-293 cells expressing FLAG-tagged full-length enthoprotin, the isolated COOH-terminal domain, or the ENTH domain. The distribution of transfected proteins was determined by anti-FLAG Western blot and compared with that of endogenous clathrin. H, homogenate; P, pellet; S, supernatant. For C and D, equal amounts of protein were loaded in each lane.
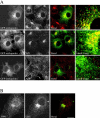
Localization of enthoprotin in COS-7 cells. (A) GFP-tagged full-length enthoprotin was transfected into COS-7 cells and its localization was compared with that of endogenous clathrin heavy chain (CHC), AP1, and AP2 determined by indirect immunofluorescence. The colocalization of enthoprotin (green) with the various markers (red) is revealed in the blended images (blend) and with higher magnification (blend mag). (B) The localization of endogenous COS-7 cell enthoprotin (red) and the clathrin heavy chain (CHC) (green) were determined by indirect immunofluorescence and the overlap of the two signals was reveled in the blended image (blend). Bar, 4 μm in the magnified panels, 20 μm in all other panels.

Enthoprotin binds to coat components of CCVs. (A) Domain models of enthoprotin GST fusion proteins. The gray bars represent the type 2–clathrin boxes (starting at amino acids 326 and 422) and the clathrin box–like sequence (starting at amino acid 342). (B and C) Various enthoprotin GST fusion proteins were coupled to glutathione-Sepharose and used in pulldown assays from brain extracts. Specifically, bound proteins were analyzed by Western blots, along with an aliquot of the brain extract (SM, starting material) with antibodies against the clathrin heavy chain (CHC), γ-adaptin, and α-adaptin.
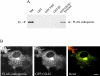
Enthoprotin binds to and colocalizes with GGA2. (A) GST fusion proteins bound to glutathione-Sepharose were used in pulldown assays from COS-7 cells expressing FLAG-tagged full-length enthoprotin. Specifically, bound proteins were analyzed by Western blotting for the FLAG epitope. (B) FLAG-tagged full-length enthoprotin was coexpressed with GFP-tagged GGA2. The enthoprotin and GGA2 fusion proteins were detected by FLAG immunostaining and GFP fluorescence, respectively. Bar, 10 μm.

Enthoprotin stimulates clathrin assembly. (A) GST or a GST fusion protein encoding the terminal domain of the clathrin heavy chain (GST-TD) were precoupled to glutathione-Sepharose beads and used in pulldowns from HEK-293 cells expressing FLAG-enthoprotin COOH-terminal or ENTH domains. Specifically bound proteins were analyzed by Western blotting for the FLAG epitope along with an aliquot of the cell lysate (SM, starting material). (B) Clathrin assembly assays were performed with increasing amounts of GST, GST–ΔC1, GST–ΔC2, and GST–ΔC3 as indicated. The GST fusion proteins and the clathrin remaining in the supernatant after high-speed centrifugation were analyzed by SDS-PAGE and Coomassie blue staining. The migratory positions of the clathrin heavy chain (CHC) and the various fusion proteins are indicated. (C) Model of potential enthoprotin function.
Similar articles
-
Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network.
Puertollano R, van der Wel NN, Greene LE, Eisenberg E, Peters PJ, Bonifacino JS. Puertollano R, et al. Mol Biol Cell. 2003 Apr;14(4):1545-57. doi: 10.1091/mbc.02-07-0109. Mol Biol Cell. 2003. PMID: 12686608 Free PMC article.
-
EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking.
Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT. Mills IG, et al. J Cell Biol. 2003 Jan 20;160(2):213-22. doi: 10.1083/jcb.200208023. Epub 2003 Jan 21. J Cell Biol. 2003. PMID: 12538641 Free PMC article.
-
Novel binding sites on clathrin and adaptors regulate distinct aspects of coat assembly.
Knuehl C, Chen CY, Manalo V, Hwang PK, Ota N, Brodsky FM. Knuehl C, et al. Traffic. 2006 Dec;7(12):1688-700. doi: 10.1111/j.1600-0854.2006.00499.x. Epub 2006 Oct 17. Traffic. 2006. PMID: 17052248
-
Peptide motifs: building the clathrin machinery.
McPherson PS, Ritter B. McPherson PS, et al. Mol Neurobiol. 2005 Aug;32(1):73-87. doi: 10.1385/MN:32:1:073. Mol Neurobiol. 2005. PMID: 16077185 Review.
-
Proteomic analysis of clathrin-coated vesicles.
McPherson PS. McPherson PS. Proteomics. 2010 Nov;10(22):4025-39. doi: 10.1002/pmic.201000253. Proteomics. 2010. PMID: 21080493 Review.
Cited by
-
Schor S, Pu S, Nicolaescu V, Azari S, Kõivomägi M, Karim M, Cassonnet P, Saul S, Neveu G, Yueh A, Demeret C, Skotheim JM, Jacob Y, Randall G, Einav S. Schor S, et al. J Biol Chem. 2022 Jun;298(6):101956. doi: 10.1016/j.jbc.2022.101956. Epub 2022 Apr 20. J Biol Chem. 2022. PMID: 35452674 Free PMC article.
-
Blondeau F, Ritter B, Allaire PD, Wasiak S, Girard M, Hussain NK, Angers A, Legendre-Guillemin V, Roy L, Boismenu D, Kearney RE, Bell AW, Bergeron JJ, McPherson PS. Blondeau F, et al. Proc Natl Acad Sci U S A. 2004 Mar 16;101(11):3833-8. doi: 10.1073/pnas.0308186101. Epub 2004 Mar 8. Proc Natl Acad Sci U S A. 2004. PMID: 15007177 Free PMC article.
-
Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin.
Lui WW, Collins BM, Hirst J, Motley A, Millar C, Schu P, Owen DJ, Robinson MS. Lui WW, et al. Mol Biol Cell. 2003 Jun;14(6):2385-98. doi: 10.1091/mbc.e02-11-0735. Epub 2003 Mar 20. Mol Biol Cell. 2003. PMID: 12808037 Free PMC article.
-
The clathrin interacting protein Clint/epsinR in rat testicular germ cells.
Redecker P. Redecker P. Histochem Cell Biol. 2005 Jun;123(4-5):457-62. doi: 10.1007/s00418-005-0777-2. Epub 2005 May 4. Histochem Cell Biol. 2005. PMID: 15875209
References
-
- Bell, A.W., M.A. Ward, W.P. Blackstock, H.N. Freeman, J.S. Choudhary, A.P. Lewis, D. Chotai, A. Fazel, J.N. Gushue, J. Paiement, et al. 2001. Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 276:5152–5165. - PubMed
-
- De Camilli, P., H. Chen, J. Hyman, E. Panepucci, A. Bateman, and A.T. Brunger. 2002. The ENTH domain. FEBS Lett. 513:11–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials