Regulation of the Escherichia coli rrnB P2 promoter - PubMed
Regulation of the Escherichia coli rrnB P2 promoter
Heath D Murray et al. J Bacteriol. 2003 Jan.
Abstract
The seven rRNA operons in Escherichia coli each contain two promoters, rrn P1 and rrn P2. Most previous studies have focused on the rrn P1 promoters. Here we report a systematic analysis of the activity and regulation of the rrnB P2 promoter in order to define the intrinsic properties of rrn P2 promoters and to understand better their contributions to rRNA synthesis when they are in their natural setting downstream of rrn P1 promoters. In contrast to the conclusions reached in some previous studies, we find that rrnB P2 is regulated: it displays clear responses to amino acid availability (stringent control), rRNA gene dose (feedback control), and changes in growth rate (growth rate-dependent control). Stringent control of rrnB P2 requires the alarmone ppGpp, but growth rate-dependent control of rrnB P2 does not require ppGpp. The rrnB P2 core promoter sequence (-37 to +7) is sufficient to serve as the target for growth rate-dependent regulation.
Figures
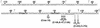
Sequence of the rrnB P2 promoter region from −112 to +21. The most frequent rrnB P2 transcription start site is designated +1. The −10 and −35 hexamers are indicated in bold, start sites are underlined, and the upstream (−112, −68, −53, and −37) or downstream (+7 and +21) endpoints used for construction of lacZ fusions are indicated by ⌈ or ⌉, respectively. The −15C insertion is indicated by the symbol ^, and substitutions are identified below the arrows.
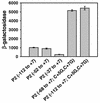
Relative activities of rrnB P2 promoter variants. β-Galactosidase activities from single-copy rrnB P2 promoter-lacZ fusions were measured in M9 minimal medium containing 0.4% glycerol as a carbon source. The rrnB P2(−68 to +7; C+5G,C+7G) fusion is the construct reported in reference . The average and standard deviation of at least three independent experiments are shown for each promoter.

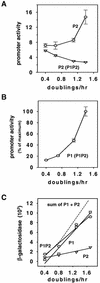
Promoter activities as a function of growth rate. β-Galactosidase activities were measured at different growth rates, obtained by growing cells in different media as described in Materials and Methods: M9 medium with 0.4% glycerol, M9 medium with 0.4% glucose, M9 medium with 0.4% glycerol plus 0.8% Casamino Acids plus tryptophan, M9 medium with 0.4% glucose plus 0.8% Casamino Acids plus tryptophan, and LB medium. Linear regressions were drawn by using SigmaPlot 5.0 (Jandel Scientific). The endpoints of the wild-type and mutant rrnB P2 promoter fragments used to construct the fusions are indicated in the panels. The lacUV5 promoter-lacZ fusion shown in panel E has been described previously (12). To enable visual comparison of the slopes, the activity of each promoter was normalized to a value of 1.0 at a growth rate of 0.9 doubling per hour (8). Strain designations and observed promoter activities (in Miller units) at a growth rate of 0.9 doubling per hour are as follows: A, RLG3851, 784 ± 34 U; B, RLG3863, 1,749 ± 72 U; C, RLG5014, 1,958 ± 236 U; D, RLG3914, 4,189 ± 252 U; E, RLG4993, 408 ± 66 U; F, RLG3915, 7,211 ± 300 U; G, RLG3898, 3,167 ± 281 U; H, RLG3897, 2,240 ± 104 U. Data from at least two independent experiments are shown for each construct.
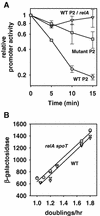
Transcription from rrnB P2 in the presence and absence of rrnB P1. (A) RNA transcribed from rrnB P2 promoter-lacZ fusions was measured directly by primer extension from lysogens grown in morpholinepropanesulfonic acid medium supplemented with 0.4% glycerol, 0.4% glucose, or 0.4% glucose plus 0.8% Casamino Acids plus tryptophan or in LB medium. Symbols: •, RLG5014, rrnB P2(−112 to +7); ▾, RLG3871, rrnB P1P2(−152 of P1 to +7 of P2). The average and standard deviation of at least three independent experiments are shown. Promoter activities are expressed in arbitrary units. (B) RNA transcribed from rrnB P1, in the context of the rrnB P1P2(−152 of P1 to +7 of P2) promoter-lacZ fusion, was measured directly by primer extension from lysogens grown in the same media used for the experiment whose results are shown in panel A. The average and standard deviation of at least three independent experiments are shown. Promoter activity is expressed as a percentage of maximum promoter activity. Absolute promoter activity should not be compared to that of rrnB P2 (see Materials and Methods). (C) β-Galactosidase activities from the same promoter-lacZ fusions described in panels A and B and from an rrnB P1-lacZ fusion. Symbols: •, RLG3871, rrnB P1P2(−152 of P1 to +7 of P2); ▪, RLG4757, rrnB P1(−152 to +50); ▾, RLG5014, rrnB P2(−112 to +7). The average of two independent experiments is shown for each promoter. The dashed line is a plot of the sum of the activities from the isolated rrnB P1-lacZ and rrnB P2-lacZ fusion constructs.
rrnB P2 is stringently controlled but does not require ppGpp for growth rate-dependent regulation. (A) RNA transcribed from rrnB P2 promoter-lacZ fusions was measured directly by primer extension following amino acid starvation induced by serine hydroxamate addition to a culture growing exponentially in LB medium (A600 of ∼0.3). The identity of the rrnB P2 promoter and the strain background are indicated for each sample. Symbols: •, RLG5014, wild-type (WT) rrnB P2(−112 to +7); ▪, RLG3987, mutant rrnB P2(−112 to +7; C−5A,A−4T,C−3A); ▾, RLG6982, wild-type rrnB P2(−112 to +7) in a relA strain. The average and standard deviation of three independent experiments are shown. (B) β-Galactosidase activity from an rrnB P2 core promoter-lacZ fusion, P2(−37 to +7), as a function of growth rate in wild-type and relA spoT mutant strains. Symbols: ▾, RLG3851, wild-type strain; •, RLG3866, relA spoT mutant strain. Data from two independent experiments are shown for each strain.
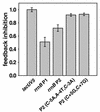
rrnB P2 promoter activity is inhibited by the presence of extra rRNA operons. Lysogens containing promoter-lacZ fusions were transformed with a multicopy plasmid containing either an intact rrnB operon (pNO1301) or an rrnB operon containing a large deletion in the 16S and 23S rRNA genes (pNO1302). Promoter activity is expressed as the ratio of the β-galactosidase activity from a promoter-lacZ fusion in a strain containing pNO1301 to that in a strain containing pNO1302 and then normalized to the value obtained from a control lacUV5 promoter as previously described (12). RLG4993, lacUV5 (−59 to +36); RLG3848, rrnB P1 (−61 to +50); RLG5014, rrnB P2(−112 to +7); RLG3987, rrnB P2(−112 to +7; C−5A,A−4T,C−3A); RLG3915, rrnB P2(−112 to +7, C+5G,C +7G). The average results and standard deviations of experiments with three independent transformants are shown.
Similar articles
-
Unique roles of the rrn P2 rRNA promoters in Escherichia coli.
Murray HD, Gourse RL. Murray HD, et al. Mol Microbiol. 2004 Jun;52(5):1375-87. doi: 10.1111/j.1365-2958.2004.04060.x. Mol Microbiol. 2004. PMID: 15165240
-
Liebig B, Wagner R. Liebig B, et al. Mol Gen Genet. 1995 Nov 27;249(3):328-35. doi: 10.1007/BF00290534. Mol Gen Genet. 1995. PMID: 7500958
-
Gafny R, Cohen S, Nachaliel N, Glaser G. Gafny R, et al. J Mol Biol. 1994 Oct 21;243(2):152-6. doi: 10.1006/jmbi.1994.1641. J Mol Biol. 1994. PMID: 7523681
-
Control of rRNA synthesis in Escherichia coli: a systems biology approach.
Dennis PP, Ehrenberg M, Bremer H. Dennis PP, et al. Microbiol Mol Biol Rev. 2004 Dec;68(4):639-68. doi: 10.1128/MMBR.68.4.639-668.2004. Microbiol Mol Biol Rev. 2004. PMID: 15590778 Free PMC article. Review.
-
Growth rate regulation in Escherichia coli.
Jin DJ, Cagliero C, Zhou YN. Jin DJ, et al. FEMS Microbiol Rev. 2012 Mar;36(2):269-87. doi: 10.1111/j.1574-6976.2011.00279.x. Epub 2011 Jun 3. FEMS Microbiol Rev. 2012. PMID: 21569058 Free PMC article. Review.
Cited by
-
Krásný L, Gourse RL. Krásný L, et al. EMBO J. 2004 Nov 10;23(22):4473-83. doi: 10.1038/sj.emboj.7600423. Epub 2004 Oct 21. EMBO J. 2004. PMID: 15496987 Free PMC article.
-
Kim CK, Lee CH, Lee SB, Oh JW. Kim CK, et al. PLoS One. 2013 Nov 4;8(11):e80109. doi: 10.1371/journal.pone.0080109. eCollection 2013. PLoS One. 2013. PMID: 24224041 Free PMC article.
-
Panlilio M, Grilli J, Tallarico G, Iuliani I, Sclavi B, Cicuta P, Cosentino Lagomarsino M. Panlilio M, et al. Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2016391118. doi: 10.1073/pnas.2016391118. Proc Natl Acad Sci U S A. 2021. PMID: 33931503 Free PMC article.
-
Farookhi H, Xia X. Farookhi H, et al. Microorganisms. 2024 Apr 10;12(4):768. doi: 10.3390/microorganisms12040768. Microorganisms. 2024. PMID: 38674712 Free PMC article.
-
Heins AL, Reyelt J, Schmidt M, Kranz H, Weuster-Botz D. Heins AL, et al. Microb Cell Fact. 2020 Jan 28;19(1):14. doi: 10.1186/s12934-020-1283-x. Microb Cell Fact. 2020. PMID: 31992282 Free PMC article.
References
-
- Afflerbach, H., O. Schroder, and R. Wagner. 1998. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol. Microbiol. 28:641-653. - PubMed
-
- Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. - PubMed
-
- Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331-345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases