Temperature-dependent conformational changes in herpes simplex virus ICP4 that affect transcription activation - PubMed
Temperature-dependent conformational changes in herpes simplex virus ICP4 that affect transcription activation
Peter Compel et al. J Virol. 2003 Mar.
Abstract
The C-terminal 500 amino acids of herpes simplex virus type 1 ICP4 are required for full activator function and viral growth and are known to participate in interactions consistent with the role of ICP4 as an activator of transcription. Oligonucleotide mutagenesis was used to target stretches of amino acids that are conserved with the ICP4 analogs of other alphaherpesviruses and were also predicted to be exposed on the surface of the molecule. Seven mutants were isolated that possessed one to three amino acid changes to the residue alanine in four regions between residues 1000 and 1200. The mutants generated were analyzed first in transfection assays and subsequently after introduction into the viral genome. A number of phenotypes representing different degrees of functional impairment were observed. In transient assays conducted at 37 degrees C, mutant M2 was indistinguishable from wild-type ICP4. Mutants M6 and M7 were marginally impaired. M3, M4, and M5 were more significantly impaired but still able to activate transcription, and M1 was completely impaired. In the context of the viral genome, M1, M3, and M7 were found to be temperature sensitive for growth. All three overproduced immediate-early (IE) proteins at the nonpermissive temperature (NPT). M3 and M7 produced early but not late proteins, and M1 produced neither early nor late proteins, at the NPT. The ICP4 proteins synthesized by all of the mutants tested were able to bind to specific ICP4 binding sites in electrophoretic mobility shift experiments. However, the DNA-protein complexes formed with the ICP4 from M1, M3, or M7 produced at the NPT possessed altered mobility. These complexes were not supershifted by a monoclonal antibody that recognizes an epitope in the C terminus; however, they were supershifted by a monoclonal antibody that recognizes the N terminus. The results suggest that the mutant forms of ICP4, while able to bind to DNA, are conformationally altered at the NPT, thus impairing the ability of the protein to activate transcription to different extents. The complete lack of ICP4 function characteristic of the M1 protein, and the inability of all the mutants to attenuate IE gene expression, suggest that the mutations additionally affect functions of the N terminus to different extents.
Figures
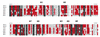
Sequence alignment of seven alphaherpesviruses corresponding to aa 1001 to 1200 of HSV-1 ICP4 and mutant loci. The entire amino acid sequences of the ICP4 analogs from HSV-1 (36), HSV-2 (19), VZV (12), pseudorabies virus (58), EHV (23), bovine herpesvirus (61), and Marek's disease virus (2) were aligned by using the Vector-NTI alignment program. Shown is the region of the alignment corresponding to the region of HSV-1 ICP4 between residues 1001 and 1200. Residues shaded in black are conserved in all seven proteins, while those shaded in red are conserved in at least four of the seven proteins. Also shown are the loci (lines above the sequence) for the M1 through M7 mutants. The line above the HSV sequence for each mutant corresponds to the sequences described in Table 1.

Expression of ICP4 from mutant plasmids. Vero cells were transfected with the indicated plasmids and were subsequently either mock infected or infected with d120 as described in Materials and Methods. Following incubation, SDS samples prepared from the cultures were subjected to SDS-PAGE, followed by Western blot analysis as described in Materials and Methods. The membranes containing the transferred SDS peptides were probed with a rabbit polyclonal antiserum raised against ICP4.
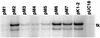
Activation of tk gene expression as a function of wt and mutant ICP4. Shown are results of a primer extension analysis for tk mRNA isolated from cells cotransfected with plasmid pLSWT and the indicated plasmid. pLSWT encodes the entire tk gene of HSV-1. The double band corresponds to the primer extension typically seen with the tk promoter/start site (29, 30).
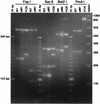
Restriction enzyme analysis of PCR-amplified products of mutant viruses and the plasmids used to generate them. DNA was isolated from cells infected with the M1, M2, M3, M6, M7, or KOS virus. The region containing the mutational loci in the DNA of the infected cell as well as that in the plasmids used to generate the mutant viruses was PCR amplified as described in Materials and Methods. The amplified products were digested with the indicated restriction enzyme and electrophoresed on agarose gels. The enzymes used to digest a mutant virus-plasmid pair were those whose sites were engineered into the plasmid by oligonucleotide mutagenesis. Shown are the EtBr-stained gel of the restriction digests and size markers for reference.
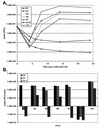
Single-step growth of mutant viruses in Vero cells. (A) Mutant viruses M1, M2, M3, M6, M7, and d120 and wt virus (KOS) were compared in a single-step growth experiment conducted at 37°C. Shown are total yields (PFU) per infected Vero cell culture as a function of time postinfection. (B) Burst sizes of mutant viruses M1, M2, M3, M6, M7, and d120 and wt virus (KOS) were compared in a single-step growth experiment conducted at 34, 37, and 39.5°C. Shown are the yields (PFU) per Vero cell for each of the viruses after 24 h of incubation at the indicated temperatures (degrees Celsius).

Synthesis of viral polypeptides in Vero cells infected with wt and ICP4-mutant viruses. Vero cells were infected with 10 PFU of the indicated viruses/cell and were incubated at the indicated temperatures. Cultures were labeled with
l-[35S]methionine from 6 to 7 h and from 12 to 13 h postinfection as described in Materials and Methods. Following the labeling period, monolayers were harvested and analyzed by SDS-PAGE as described in Materials and Methods. Shown are autoradiographic images of infected cell polypeptides. Positions of representative infected cell polypeptides are indicated by arrows and numbers on the right (for example, “4” stands for ICP4).
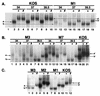
DNA binding of mutant ICP4 proteins. EMSA was used to observe the abilities of the mutant ICP4 proteins to bind to a specific ICP4 binding site. Details of the EMSA are provided in Materials and Methods. In some reactions the ICP4-specific monoclonal antibody 58S (+) or 1101 (#) was used to supershift complexes formed by ICP4 protein. Designations: a, mobility of ICP4-DNA complexes formed with extracts from KOS-infected cells; a′, mobility to which these complexes were supershifted by antibodies 58S and 1101; b, mobility (greater than a) of some ICP4-DNA complexes formed with extracts from M1-infected cells; b′, mobility to which “b” complexes were supershifted by 1101. (A) Comparison of DNA-protein complexes formed with KOS and M1 extracts prepared from cells infected at 34, 37, and 39.5°C. Shown are autoradiographic images of the region of the gel containing the ICP4-DNA protein complexes. Lane 1 represents the DNA-protein complexes in this region of the gel formed by using extracts from d120-infected cells. (B) Comparison of DNA-protein complexes formed with M3, M7, and KOS extracts prepared from cells infected at 34, 37, and 39.5°C. For KOS, only the 37°C samples are shown. (C) Comparison of DNA-protein complexes formed with M3, M2, M1, and KOS ICP4s purified from cells infected at 37°C.

Western blot analysis of mutant ICP4 proteins using different antibody preparations. Three-microliter portions of the protein extracts that were also used in the EMSA experiments (Fig. 7) were combined with 3 μl of Laemmli sample buffer, boiled for 3 min, and subsequently resolved by SDS-PAGE (6% polyacrylamide). The SDS-peptides were transferred to polyvinylidene difluoride membranes for Western blot analysis as described in Materials and Methods. Three different antibodies were used to probe the membranes: N15 (rabbit polyclonal), 58S (mouse monoclonal), and 1101 (mouse monoclonal).

Loci for known ts mutants in the C terminus of ICP4. (Top) The region of ICP4 expressed from mutant n208 (17) and its phenotype with respect to activation of early genes and autoregulation are shown above a scale representing the entire 1,298 aa of HSV-1 ICP4. (Bottom) The loci of known ts mutants of strains KOS and 17 that are defective and permissive for early gene expression are shown above and below a scale representing aa 800 to 1298, respectively. Loci for defective strain 17 mutants tsD and tsT (48), defective KOS mutants tsB2, tsB21, tsB27, tsB28, and tsB32 (18), permissive strain 17 mutants ts1221, ts1219, ts1225, ts1211, and ts1223 (44), and permissive KOS mutants ts48 and ts303 (13) are represented as described previously. Those for M1, M3, and M7 are from this study.
Similar articles
-
Wagner LM, Lester JT, Sivrich FL, DeLuca NA. Wagner LM, et al. J Virol. 2012 Jun;86(12):6862-74. doi: 10.1128/JVI.00651-12. Epub 2012 Apr 11. J Virol. 2012. PMID: 22496239 Free PMC article.
-
Samaniego LA, Webb AL, DeLuca NA. Samaniego LA, et al. J Virol. 1995 Sep;69(9):5705-15. doi: 10.1128/JVI.69.9.5705-5715.1995. J Virol. 1995. PMID: 7637016 Free PMC article.
-
Zhu Z, Schaffer PA. Zhu Z, et al. J Virol. 1995 Jan;69(1):49-59. doi: 10.1128/JVI.69.1.49-59.1995. J Virol. 1995. PMID: 7983745 Free PMC article.
-
Zhu Z, Cai W, Schaffer PA. Zhu Z, et al. J Virol. 1994 May;68(5):3027-40. doi: 10.1128/JVI.68.5.3027-3040.1994. J Virol. 1994. PMID: 8151771 Free PMC article.
Cited by
-
Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1.
Ferenczy MW, Ranayhossaini DJ, Deluca NA. Ferenczy MW, et al. J Virol. 2011 May;85(10):4993-5002. doi: 10.1128/JVI.02265-10. Epub 2011 Mar 16. J Virol. 2011. PMID: 21411540 Free PMC article.
-
Pinnoji RC, Bedadala GR, George B, Holland TC, Hill JM, Hsia SC. Pinnoji RC, et al. Virol J. 2007 Jun 7;4:56. doi: 10.1186/1743-422X-4-56. Virol J. 2007. PMID: 17555596 Free PMC article.
-
Wagner LM, Bayer A, Deluca NA. Wagner LM, et al. J Virol. 2013 Jan;87(2):1010-8. doi: 10.1128/JVI.02844-12. Epub 2012 Nov 7. J Virol. 2013. PMID: 23135715 Free PMC article.
-
Petrik DT, Schmitt KP, Stinski MF. Petrik DT, et al. J Virol. 2007 Jun;81(11):5807-18. doi: 10.1128/JVI.02437-06. Epub 2007 Mar 21. J Virol. 2007. PMID: 17376893 Free PMC article.
-
Zabierowski S, DeLuca NA. Zabierowski S, et al. J Virol. 2004 Jun;78(12):6162-70. doi: 10.1128/JVI.78.12.6162-6170.2004. J Virol. 2004. PMID: 15163709 Free PMC article.
References
-
- Alwine, J. C., W. L. Steinhart, and C. W. Hill. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60:302-307. - PubMed
-
- Anderson, A. S., A. Francesconi, and R. W. Morgan. 1992. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology 189:657-667. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources