ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation - PubMed
- ️Wed Jan 01 2003
ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation
Kathryn S Sciabica et al. EMBO J. 2003.
Abstract
Infection with some viruses can alter cellular mRNA processing to favor viral gene expression. We present evidence that herpes simplex virus 1 (HSV-1) protein ICP27, which contributes to host shut-off by inhibiting pre-mRNA splicing, interacts with essential splicing factors termed SR proteins and affects their phosphorylation. During HSV-1 infection, phosphorylation of several SR proteins was reduced and this correlated with a subnuclear redistribution. Exogenous SR proteins restored splicing in ICP27-inhibited nuclear extracts and SR proteins isolated from HSV-1-infected cells activated splicing in uninfected S100 extracts, indicating that inhibition occurs by a reversible mechanism. Spliceosome assembly was blocked at the pre-spliceosomal complex A stage. Furthermore, we show that ICP27 interacts with SRPK1 and relocalizes it to the nucleus; moreover, SRPK1 activity was altered in the presence of ICP27 in vitro. We propose that ICP27 modifies SRPK1 activity resulting in hypophosphorylation of SR proteins impairing their ability to function in spliceosome assembly.
Figures
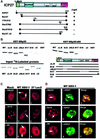
Fig. 1. ICP27 interacts with SRp20. (A) The ICP27 coding region is shown schematically, including a leucine-rich sequence (L-rich), NLS, RNA- binding region (RGG box), three putative KH domains and a zinc finger-like motif (CCHC). ICP27 mutations fused to the Gal4 DNA-binding domain were tested for interaction with SRp20 fused to the Gal4 activation domain as measured by β-galactosidase production. (B) GST-binding assays were performed with GST–SRp20 (left panel) or GST–SRp20ΔRS (right panel) and in vitro-translated WT ICP27 and mutants ΔL-R, D2ΔS5, ΔNLS, H17, S18 and ΔC. Input 35S-labeled proteins are shown (lower left panel) as is a schematic diagram of the mutants (lower right panel). (C) HeLa cells that were mock-infected (panels 1, 4 and 7), infected with WT HSV-1 (panels 2, 5 and 8) or 27-LacZ (panels 3, 6 and 9) for 3 h were stained with anti-SRp20 (panels 1–3), anti-SC35 (panels 4–6) or anti-SR (panels 7–9). Arrows indicate examples of coalesced SR proteins. (D) HSV-1-infected cells were double labeled with the indicated SR antibodies (red) and anti-ICP27 (green). The fields were merged to show co-localization (arrows) in yellow (panels 3, 6 and 9).

Fig. 2. ICP27 interacts with SR proteins in infected cells. (A) A nuclear extract from HSV-1-infected cells labeled with 32Pi from 1–6 h after infection was immunoprecipitated with anti-ICP27 antibody (left panels) or mAB104 (right panels). Phospho-labeled proteins were detected by autoradiography. ICP27 proteolytic degradation products are marked with arrowheads. The blots were probed with mAb104 (left) or anti-ICP27 (right). SR protein bands are indicated. Asterisks mark non-specific bands that reacted with the secondary antibody. Plus signs mark heavy and light chain IgG from the immunoprecipitation. (B) 293 cells transfected with pFlag-SRp20 were mock-infected or infected as indicated. Extracts were immunoprecipitated with anti-SRp20 antibody and the blot was probed with anti-ICP27 antibody. (C) In vitro binding was performed by mixing extracts from cells transfected with pFlag-SRp20 or pCMV-ICP27. Extracts were treated (+) or not (–) with RNase and immunoprecipitated with anti-Flag antibody. The blot was probed with anti-ICP27 antibody. (D) Extracts from cells transfected with pFlag-SRp20ΔRS or pCMV-ICP27 were mixed and immunoprecipitated with anti-ICP27 or anti-Flag antibody as indicated. The blots were probed with anti-ICP27 or anti-Flag antibody. In NE lanes, a portion of each nuclear extract was fractionated without immunoprecipitation. Asterisks mark heavy and light chain IgG. A bracket marks ICP27 degradation products (NE and lanes 1 and 2).

Fig. 3. ICP27 alters SR phosphorylation. (A) Nuclear extracts were prepared from 293 cells transfected with pFlag-SRp20, then mock- or HSV- infected for 4 and 5 h, and labeled with 32Pi. Extracts were immunoprecipitated with anti-Flag antibody. Radiolabeled proteins are shown in the upper panel. The blot was probed with anti-Flag antibody (lower panel). The plus sign marks IgG light chain from the immunoprecipitation. (B) 293 cells transfected with pFlag-SRp20 (left) or pFlag-SRp40 (right) were infected as indicated and labeled with 32Pi from 1 to 6 h after infection. Extracts were fractionated directly by SDS–PAGE. Radiolabeled proteins (upper panels) and western blot analysis with anti-Flag antibody (lower panels) are shown. (C) Nuclear extracts were prepared from 293 cells co-transfected with pFlag-SRp40 and plasmids expressing WT ICP27, S5, N2, or S18 and labeled with 32Pi for 5 h beginning 20 h after transfection. Extracts were immunoprecipitated with anti-Flag (left) or anti-ICP27 (middle). Radiolabeled proteins (upper panels) and western blot analysis with anti-Flag or anti-ICP27 (lower middle panels) are shown. Extracts containing Flag-SRp40 were mixed with WT ICP27, N2 or S18 and immunoprecipitated with anti-Flag antibody. The blot was probed with anti-ICP27 (right panel). (D) 293 cells were transfected with pFlag-SRp20. Uninfected (UN) and WT HSV-1-infected cells were labeled with 32Pi for 5 h. Nuclear extracts were fractionated directly (left panel) and the blot was later probed with anti-ICP27 and anti-Flag antibody (middle panels), or were immunoprecipitated with anti-ICP27 or anti-Flag antibody (right panels). (E) [γ-32P]ATP was added to mock or HSV-1 nuclear splicing extracts, each containing 60 µg of protein, and incubated for 3 h at 30°C. SR proteins were precipitated with MgCl2 and fractionated by SDS–PAGE.
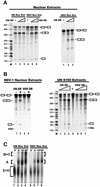
Fig. 4. SR proteins restore splicing to HSV-inhibited extracts. (A) 32P-labeled β-globin pre-mRNA was added to nuclear splicing extracts from uninfected (left panel, lanes 1–3) or HSV-infected (lanes 4–6) cells. Splicing extracts were not supplemented (lanes 1 and 4) or were supplemented with 200 ng (lanes 2 and 5) or 400 ng (lanes 3 and 6) of SR proteins from uninfected HeLa cells. AdML pre-mRNA was added to HSV nuclear splicing extracts (right panel, lanes 2 and 3). SR proteins from uninfected HeLa cells were added at 200 ng (lane 2) or 400 ng (lanes 3). (B) Purified SR proteins (200 ng) from uninfected (left panel, lanes 1 and 2) or HSV-1-infected (lanes 3 and 4) cells were added to nuclear splicing extracts from HSV-1-infected cells to which β-globin pre-mRNA was added. S100 extracts from uninfected cells were supplemented with 200, 400 or 800 ng of SR proteins from uninfected cells (right panel, lanes 1–3) or with the same amounts of SR proteins from HSV-1-infected cells (lanes 5–7). SR proteins were not added to lane 4. Splicing of β-globin pre-mRNA was monitored. (C) AdML pre-mRNA was added to splicing extracts from uninfected and HSV-1-infected HeLa cells that were analyzed for spliceosome complex formation on 4% non-denaturing polyacrylamide gels. Spliceosome assembly complexes A, B and C are labeled. The early complex (E) and hnRNP–pre-mRNA complex (H) co-migrated as a diffuse band labeled E+H.

Fig. 5. ICP27 interacts with SRPK1. (A) Cells were transfected with pFlag-SRPK1 (lanes 2–5) or a control plasmid (lanes 1, 6 and 7) and extracts were prepared from transfected cells that were mock-infected (lanes 2 and 7), infected with WT HSV-1 (lanes 1, 3, 5 and 6) or 27-LacZ (lane 4) and labeled with 32Pi from 1 to 5 h after infection. Immunoprecipitation was performed with anti-Flag (lanes 1–4) or anti-ICP27 (lanes 5–7). Radiolabeled proteins (left) and immunoblot analysis with anti-Flag antibody (right) are shown. (B) Extracts from cells transfected with pFlag-SRPK1 or pCMV-ICP27 were treated with RNase (+) or buffer (–) and then mixed. Immunoprecipitation was performed with anti-Flag and the blot was probed with anti-ICP27 antibody. (C) In vitro binding assays were performed with His-SRPK1 and in vitro-translated WT ICP27 or mutant proteins as indicated. Input [35S]methionine-labeled proteins and a schematic showing the positions of the mutations are shown. (D) Cells were co-transfected with Flag-Srp40 and plasmids expressing WT ICP27, D2ΔS5, S18 or Δ63–100. Nuclear extracts were fractionated and western blot analysis was performed with anti-SR antibody (upper left panel), anti-Flag (lower left panel) and anti-ICP27 (right panel).
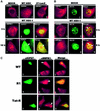
Fig. 6. ICP27 relocalizes SRPK1 to the nucleus. (A) Cells that were mock-, WT HSV-1- or 27-LacZ-infected for 3 h were stained with anti-SRPK1 antibody (panels 1–3). Double labeling with anti-SRPK1 (red) and anti-ICP27 (green) was performed on cells infected with WT HSV-1 for 3 (panels 4–6) and 10 h (panels 7–9). Merged images show co-localization in yellow (panels 6 and 9). ICP27 perinuclear staining is marked by an arrow (panel 8), as is SRPK1 nuclear staining (panel 9). (B) Double labeling with anti-SRPK1 (panels 1, 4 and 7) and anti-SC35 (panels 2, 5 and 8) was performed on cells that were mock-infected (panels 1–3) or infected with HSV-1 for 3 (panels 4–6) and 8 h (panels 7–9). Areas of co-localization (yellow) in the merged images are denoted by arrows. Asterisks mark regions of SC35 staining that do not contain SRPK1. (C) Cells were transfected with pCMV-ICP27, R1 or Tat-R. Double labeling was performed with anti-ICP27 (panels 1, 4 and 7) and anti-SRPK1 (panels 2, 5 and 8). The merged images are shown (lanes 3, 6 and 9).

Fig. 7. ICP27 alters SRPK1 activity in vitro. (A) His-SRPK1 was pre-incubated with immunoprecipitated ICP27 or control antibody–protein A– Sepharose complexes. SR proteins purified from HeLa cells (lanes 3 and 4), GST–SF2/ASF (lanes 5 and 6) and His-SC35 (lanes 7 and 8) were added as was [γ-32P]ATP. Heavy and light chain IgG from antibody complexes are marked (+) in the Coomassie Blue-stained gel (left). (B) The SRPK1 homolog Sky1p site in Np13p is shown with putative SRPK1 sites in ICP27. (C) SRPK1, immunoprecipitated ICP27 or control antibody complexes were pre-incubated and SRp30 proteins from HeLa cells were added to kinase reactions. The bracket marks the hypophosphorylated species. Immunoblot analysis with anti-ICP27 antibody is shown below. (D) Kinase assays were performed with His-SRPK1 and immunoprecipitated WT ICP27 or mutants D2ΔS5, Δ63–100, S5 and ΔNLS with GST–SF2/ASF as the substrate. The Coomassie Blue-stained gels are shown below.
Similar articles
-
Molecular Mechanism of SR Protein Kinase 1 Inhibition by the Herpes Virus Protein ICP27.
Tunnicliffe RB, Hu WK, Wu MY, Levy C, Mould AP, McKenzie EA, Sandri-Goldin RM, Golovanov AP. Tunnicliffe RB, et al. mBio. 2019 Oct 22;10(5):e02551-19. doi: 10.1128/mBio.02551-19. mBio. 2019. PMID: 31641093 Free PMC article.
-
Lindberg A, Kreivi JP. Lindberg A, et al. Virology. 2002 Mar 1;294(1):189-98. doi: 10.1006/viro.2001.1301. Virology. 2002. PMID: 11886277
-
Hardy WR, Sandri-Goldin RM. Hardy WR, et al. J Virol. 1994 Dec;68(12):7790-9. doi: 10.1128/JVI.68.12.7790-7799.1994. J Virol. 1994. PMID: 7966568 Free PMC article.
-
Properties of an HSV-1 regulatory protein that appears to impair host cell splicing.
Sandri-Goldin RM. Sandri-Goldin RM. Infect Agents Dis. 1994 Apr-Jun;3(2-3):59-67. Infect Agents Dis. 1994. PMID: 7812656 Review.
-
The many roles of the regulatory protein ICP27 during herpes simplex virus infection.
Sandri-Goldin RM. Sandri-Goldin RM. Front Biosci. 2008 May 1;13:5241-56. doi: 10.2741/3078. Front Biosci. 2008. PMID: 18508584 Review.
Cited by
-
Johnson KE, Knipe DM. Johnson KE, et al. Virology. 2010 Jan 5;396(1):21-9. doi: 10.1016/j.virol.2009.09.021. Epub 2009 Oct 31. Virology. 2010. PMID: 19879619 Free PMC article.
-
Park R, Miller G. Park R, et al. J Virol. 2018 Sep 26;92(20):e01254-18. doi: 10.1128/JVI.01254-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068640 Free PMC article.
-
Rice BL, Lochmann TL, Parent LJ. Rice BL, et al. Viruses. 2020 Mar 27;12(4):370. doi: 10.3390/v12040370. Viruses. 2020. PMID: 32230826 Free PMC article.
-
Sanders LS 3rd, Comar CE, Srinivas KP, Lalli J, Salnikov M, Lengyel J, Southern P, Mohr I, Wilson AC, Rice SA. Sanders LS 3rd, et al. J Virol. 2023 Jul 27;97(7):e0195722. doi: 10.1128/jvi.01957-22. Epub 2023 Jun 13. J Virol. 2023. PMID: 37310267 Free PMC article.
-
Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Walsh CM, et al. J Biol Chem. 2013 Jan 25;288(4):2816-28. doi: 10.1074/jbc.M112.410803. Epub 2012 Dec 11. J Biol Chem. 2013. PMID: 23233666 Free PMC article.
References
-
- Champion-Arnaud P. and Reed,R. (1994) The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev., 8, 1974–1983. - PubMed
-
- Fu X.D. and Maniatis,T. (1990) Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature, 343, 437–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous