Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis - PubMed
- ️Sat Nov 24 2164
Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis
Shanfa Lu et al. Plant Cell. 2005 Aug.
Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that can play crucial regulatory roles in eukaryotes by targeting mRNAs for silencing. To test whether miRNAs play roles in the regulation of wood development in tree species, we isolated small RNAs from the developing xylem of Populus trichocarpa stems and cloned 22 miRNAs. They are the founding members of 21 miRNA gene families for 48 miRNA sequences, represented by 98 loci in the Populus genome. A majority of these miRNAs were predicted to target developmental- and stress/defense-related genes and possible functions associated with the biosynthesis of cell wall metabolites. Of the 21 P. trichocarpa miRNA families, 11 have sequence conservation in Arabidopsis thaliana but exhibited species-specific developmental expression patterns, suggesting that even conserved miRNAs may have different regulatory roles in different species. Most unexpectedly, the remaining 10 miRNAs, for which 17 predicted targets were experimentally validated in vivo, are absent from the Arabidopsis genome, suggesting possible roles in tree-specific processes. In fact, the expression of a majority of the cloned miRNAs was upregulated or downregulated in woody stems in a manner consistent with tree-specific corrective growth against tension and compression stresses, two constant mechanical loads in trees. Our results show that plant miRNAs can be induced by mechanical stress and may function in one of the most critical defense systems for structural and mechanical fitness.
Figures

Predicted Stem-Loop Structures of Precursors Containing the Newly Cloned miRNA Sequence (Red).
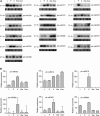
Developmental and Mechanical Stress–Responsive Expression of Populus miRNAs. (A) RNA gel blots of total RNA isolated from leaf (L), phloem (P), developing xylem (X), tension-stressed developing xylem (Xtw), and compression-stressed developing xylem (Xow) were probed with end-labeled oligonucleotides. The 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls. nt, nucleotides. (B) Relative expression of miRNAs in leaf (L), phloem (P), developing xylem (X), tension-stressed developing xylem (Xtw), and compression-stressed developing xylem (Xow) was analyzed by real-time PCR. The 5.8S rRNA was selected as a reference. The data are means of three measurements ±
se.

Experimental Validation of the Predicted mRNA Targets for the Newly Cloned ptr-miR408, 473a, 474a, 475a, and 477a. The mRNA cleavage sites were determined by modified 5′ RNA ligase-mediated RACE. Heavy black lines represent ORFs. The lines flanking gray regions represent nontranslated regions (if known) and miRNA complementary sites, with the nucleotide positions within the ORF indicated. The mRNA sequence of each complementary site from 5′ to 3′ and the cloned miRNA sequence (bold) from 3′ to 5′ are shown in the expanded regions. Watson-Crick pairing (vertical dashes) and G:U wobble pairing (circles) are indicated. Vertical arrows indicate the 5′ termini of miRNA-guided cleavage products, as identified by 5′-RACE, with the frequency of clones shown. Positions of the nesting and the nested primers used for 5′-RACE are indicated by horizontal arrows. For targets eugene3.03440011, eugene3.00700255, and eugene3.08910001 predicted for ptr-miR474a, 5′-RACE was performed using the same set of primers, yielding products with the identical sequence for which origin could not be certain. Therefore, at least one of them is an authentic target for ptr-miR474a.

Experimental Validation of the Predicted mRNA Targets for the Newly Cloned ptr-miR475a, 476a, 478a, 479, 480a, and 482. The ptr-miR475a targets shown in this figure and in Figure 3 were validated using the same set of degenerate primers. The partial flanking sequence of ptr-miR482 in the precursor is also shown.
Similar articles
-
MicroRNAs expression patterns in the response of poplar woody root to bending stress.
Rossi M, Trupiano D, Tamburro M, Ripabelli G, Montagnoli A, Chiatante D, Scippa GS. Rossi M, et al. Planta. 2015 Jul;242(1):339-51. doi: 10.1007/s00425-015-2311-7. Epub 2015 May 12. Planta. 2015. PMID: 25963516
-
Stress-responsive microRNAs in Populus.
Lu S, Sun YH, Chiang VL. Lu S, et al. Plant J. 2008 Jul;55(1):131-51. doi: 10.1111/j.1365-313X.2008.03497.x. Plant J. 2008. PMID: 18363789
-
Conservation and divergence of microRNAs in Populus.
Barakat A, Wall PK, Diloreto S, Depamphilis CW, Carlson JE. Barakat A, et al. BMC Genomics. 2007 Dec 31;8:481. doi: 10.1186/1471-2164-8-481. BMC Genomics. 2007. PMID: 18166134 Free PMC article.
-
Plant and animal microRNAs: similarities and differences.
Millar AA, Waterhouse PM. Millar AA, et al. Funct Integr Genomics. 2005 Jul;5(3):129-35. doi: 10.1007/s10142-005-0145-2. Epub 2005 May 5. Funct Integr Genomics. 2005. PMID: 15875226 Review.
-
Douglas CJ, Ehlting J. Douglas CJ, et al. Transgenic Res. 2005 Oct;14(5):551-61. doi: 10.1007/s11248-005-8926-x. Transgenic Res. 2005. PMID: 16245146 Review.
Cited by
-
Fan T, Li X, Yang W, Xia K, Ouyang J, Zhang M. Fan T, et al. PLoS One. 2015 May 29;10(5):e0125833. doi: 10.1371/journal.pone.0125833. eCollection 2015. PLoS One. 2015. PMID: 26023934 Free PMC article.
-
The MicroRNAs and their targets in the channel catfish (Ictalurus punctatus).
Barozai MY. Barozai MY. Mol Biol Rep. 2012 Sep;39(9):8867-72. doi: 10.1007/s11033-012-1753-2. Epub 2012 Jun 24. Mol Biol Rep. 2012. PMID: 22729904
-
Zhang Y, Wang W, Chen J, Liu J, Xia M, Shen F. Zhang Y, et al. Int J Mol Sci. 2015 Jun 30;16(7):14749-68. doi: 10.3390/ijms160714749. Int J Mol Sci. 2015. PMID: 26133244 Free PMC article.
-
Zhou H, Li C, Qiu X, Lu S. Zhou H, et al. Plants (Basel). 2019 Nov 11;8(11):490. doi: 10.3390/plants8110490. Plants (Basel). 2019. PMID: 31717988 Free PMC article.
-
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X. Xu Q, et al. BMC Genomics. 2010 Apr 17;11:246. doi: 10.1186/1471-2164-11-246. BMC Genomics. 2010. PMID: 20398412 Free PMC article.
References
-
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. - PubMed
NOTE ADDED IN PROOF
-
- miRNA and miRNA targets were independently annotated by Matthew Jones-Rhoades (Whitehead Institute for Biomedical Research; http://web.wi.mit.edu/bartel/pub/) as part of the assembly and annotation efforts of the International Populus Genome Consortium (http://www.ornl.gov.sci/ipgc/).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources