A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement - PubMed
A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement
Yun Liu et al. Immunity. 2007 Oct.
Abstract
V(D)J recombination is initiated by the recombination activating gene (RAG) proteins RAG-1 and RAG-2. The ability of antigen-receptor-gene segments to undergo V(D)J recombination is correlated with spatially- and temporally-restricted chromatin modifications. We have found that RAG-2 bound specifically to histone H3 and that this binding was absolutely dependent on dimethylation or trimethylation at lysine 4 (H3K4me2 or H3K4me3). The interaction required a noncanonical plant homeodomain (PHD) that had previously been described within the noncore region of RAG-2. Binding of the RAG-2 PHD finger to chromatin across the IgH D-J(H)-C locus showed a strong correlation with the distribution of trimethylated histone H3 K4. Mutation of a conserved tryptophan residue in the RAG-2 PHD finger abolished binding to H3K4me3 and greatly impaired recombination of extrachromosomal and endogenous immunoglobulin gene segments. Together, these findings are consistent with the interpretation that recognition of hypermethylated histone H3 K4 promotes efficient V(D)J recombination in vivo.
Figures
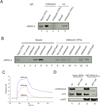
Specific binding of RAG-2 to histone H3 containing di- or trimethylated lysine 4. (A) Whole-cell lysates of 293T cells expressing full-length, wild-type RAG-2 were incubated with streptavidin bead-bound, biotinylated peptides corresponding to residues 1 – 21 of histone H3. Peptides were unmodified (H3) or trimethylated at lysine 4 (H3K4me3). Bead-bound protein was fractionated by SDS-PAGE and RAG-2 was detected by immunoblotting. Portions (10%) of the lysate (Input) or unbound protein were assayed in parallel. (B) Lysates of 293T cells expressing wild-type RAG-2 were incubated with unmodified or modified histone H3 peptides, affixed to beads. Modifications are indicated above. All peptides correspond to residues 1 – 21 of histone H3 except for the H3K27me3 peptide, which corresponds to residues 21 – 44. Bound protein was fractionated as in (A) and RAG-2 was detected by immunobloting. Portions (10%) of the lysate (Input) or unbound protein were assayed in parallel. (C) Surface plasmon resonance binding curves for GST-RAG-2PHD association with histone H3 peptides as a function of lysine 4 methylation. Biotin-tagged H3 peptides (residues 1 – 21) were immobilized at 0.06 ng each on flow cells of a sensor chip. GST-RAG-2PHD (5 µM) was injected (solid arrow) for 2 min, followed by injection of buffer alone (open arrow). Binding curves were deduced after subtracting the response of the reference surface. (D) Methylation at lysine 4 is required for binding of the RAG-2 PHD finger to intact histone H3 in vitro. GST-RAG-2PHD was immobilized on glutathione-coated beads and incubated with histones isolated from wild-type or set1-null S. cerevisiae. Histone H3 trimethylated at lysine 4, bulk H3 and GST were detected in input and bound fractions by immunoblotting.

Binding of RAG-2 to tri-Me H3K4 is mediated by the PHD finger. (A) Representation of proteins used in binding assays. All constructs carry an N-terminal GST tag. Full-length RAG-2 is shown at top; vertical lines indicate the C-terminal end of the core domain (387), the boundaries of the PHD domain (419 – 481) and the positions of amino acid substitutions (W453A, C478A). (B) The RAG-2 PHD finger mediates direct binding to tri-Me H3K4. Proteins described in (A) were expressed in E.coli, purified on glutathione-agarose and assayed for binding to biotinylated H3K4me3 peptide immobilized on streptavidin-linked beads. Bound protein (left) or 10% input protein (right) was fractionated by SDS-PAGE and detected by Coomassie blue. (C) Lysates of mock transfected 293T cells or cells expressing full-length wild-type RAG-2 or RAG-2(W453A) were assayed for binding to the biotinylated H3K4me3 peptide as above (lanes 4 – 6). Bound protein was fractionated by SDS-PAGE and detected by immunoblotting for RAG-2. Input protein (10%) was analyzed in parallel (lanes 1 – 3). (D) RAG-2(W453A) retains the ability to associate with RAG-1. Lysates of 293T cells coexpressing a c-myc-tagged MBP-RAG-1 fusion and RAG-2 or RAG-2(W453A) were adsorbed to amylose beads. Bound protein (lanes 6 – 10) was fractionated by SDS PAGE and detected by immunoblotting for c-myc (top) or RAG-2 (bottom). Input protein (10%) was assayed in parallel (lanes 1 – 5).
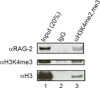
Pro-B cell lysates contain RAG-2 bound to histone H3 di- or trimethylated at lysine 4. Bone marrow cells were cultured in IL-7 for 10 d to generate B220+CD43+ pro-B cells. Protein was immunoprecipitated from pro-B cell lysates with an antibody specific for H3K4me2 and H3K4me3 or with an isotype control. Immunoprecipitates (lanes 2 and 3) were fractionated by SDS-PAGE and assayed by immunoblotting for RAG-2 (top), H3K4me3 (middle) and histone H3 (bottom). Input protein (20%) was assayed as a reference (lane 1).

Binding of the RAG-2 PHD finger to chromatin in the IgH D–JH-C region is correlated with the density of trimethylated histone H3 K4. (A) Schematic representation of the IgH D–JH-C region through the Cγ3 exons. Gray boxes, D gene segments. Black vertical lines, JH gene segments; open boxes, Cμ, Cδ and Cγ3 exons. Black oval, Eμ intronic enhancer. Black dots below indicate the approximate locations of primer pairs for the segments indicated. (B) Precipitation of chromatin from the RAG-2-deficient 63-12 cell line with an anti-H3K4me2 antibody (left), an anti-H3K4me3 (middle) antibody, wild-type GST-RAG-2PHD (right, open bars) or the mutant GST-RAG-2PHD(W453A) (right, filled bars). DNA recovered from chromatin precipitates was analyzed by quantitative real-time PCR (qRT-PCR). The fold enrichment of each amplified DNA fragment, indicated below as in (A), was determined relative to input DNA and then normalized to that of α-actin. The promoter region of γ-actin was amplified as a positive control. Error bars indicate standard deviations (n = 3).

A mutation abolishing binding of H3K4me3 by the RAG-2 PHD finger impairs recombination of endogenous immunoglobulin gene segments in pro-B cells. (A) RAG-2(W453A) retains catalytic activity in vitro. Core RAG-1 (cR1) and complexes of core RAG-1 with core RAG-2 (cR2), full-length RAG-2 (fR2) or full-length RAG-2(W453A) (fR2[WA]) were assayed in vitro for cleavage of 12-RSS (lanes 1 – 5) or 23-RSS (lanes 6 – 10) substrates; (−), no protein added. Positions of nicked and hairpin products are indicated. (B) EGFP expression in infected cells. RAG-2−/− R2FL63-12 pro-B cells were infected with control lentivirus or with a lentivirus expressing wild-type RAG-2 or RAG-2(W453A). Infection was confirmed by flow cytometric detection of EGFP, which was encoded by all three lentiviruses, at 2 d after infection. Numbers indicate percentages of cells in gated areas. RAG-2 mRNA was assayed by qRT-PCR amplification of reverse transcripts from total RNA isolated 8 d after infection. The amounts of RAG-2 mRNA in cells infected with control, RAG-2 or RAG-2(W453A) lentiviruses, relative to γ-actin mRNA, were 0.00, 1.00 and 3.47, respectively. (C) Detection of completed D-to-JH rearrangements. D-to-JH rearrangements were assayed 8 d after infection by PCR using genomic DNA from control- or RAG-2-infected pro-B cells as template as indicated above. Amplification was performed in the absence of template in parallel (- template). Forward primers were specific for DFL16.1 (top panel) or DSP2 (middle panel); the reverse primer initiates synthesis 3’ of JH4. Products were separated by gel electrophoresis and detected by hybridization to a radiolabeled, locus-specific probe that recognizes all JH segments. Genomic DNA from C57BL/6 mouse spleen was used as a positive control for D-to-JH rearrangements. The β-globin locus (bottom panel) was amplified as a control for gel loading.
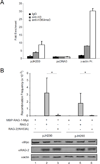
A mutation abolishing binding of H3K4me3 by the RAG-2 PHD finger impairs recombination of extrachromosomal substrates. (A) Association of pJH200 with histone H3 and H3K4me3 in NIH3T3 cells. Chromatin was precipitated from transfected cells with control IgG (filled bar), an anti-H3 antibody (shaded bar) or an anti-H3K4me3 antibody (open bar) and detected by qRT-PCR. Plasmid pJH200 was detected by amplification across the 12-spacer RSS; the cotransfected, non-replicative plasmid pcDNA3 was assayed by amplification of neo. The endogenous promoter region of γ-actin was amplified as a positive control. The fold enrichment of each amplified DNA fragment was determined relative to input DNA and then normalized to that of the f1 origin region of pcDNA3. Error bars indicate standard deviations (n = 2). (B) Plasmids encoding a c-myc-tagged MBP-RAG-1 fusion and RAG-2 or RAG-2(W453A) were transiently cotransfected with the recombination substrate pJH200 or pJH290 into NIH3T3 cells. In control experiments plasmid pcDNA3 was transfected instead of the RAG-2 encoding plasmid. Plasmids were introduced in the combinations indicated. At 48 hr after transfection plasmids were recovered and transfected into E. coli; transfectants were scored for resistance to ampicillin (Ampr) or ampicillin and chloramphenicol (Ampr + Camr). Recombination frequency (mean ± S.D., n = 2) was calculated as the ratio of double to single resistant colonies (Ampr + Camr/Ampr). (*), P < 0.04, two-tailed t-test. Cell lysates were analyzed by immunoblotting for RAG-1 (c-myc), RAG-2 or actin as indicated.
Comment in
-
Targeting V(D)J recombinase: putting a PHD to work.
Oltz EM, Osipovich O. Oltz EM, et al. Immunity. 2007 Oct;27(4):539-41. doi: 10.1016/j.immuni.2007.10.001. Immunity. 2007. PMID: 17967406 Review.
Similar articles
-
May MR, Bettridge JT, Desiderio S. May MR, et al. J Biol Chem. 2020 Jul 3;295(27):9052-9060. doi: 10.1074/jbc.RA120.014382. Epub 2020 May 15. J Biol Chem. 2020. PMID: 32414844 Free PMC article.
-
RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination.
Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. Matthews AG, et al. Nature. 2007 Dec 13;450(7172):1106-10. doi: 10.1038/nature06431. Epub 2007 Nov 21. Nature. 2007. PMID: 18033247 Free PMC article.
-
Lu C, Ward A, Bettridge J, Liu Y, Desiderio S. Lu C, et al. Cell Rep. 2015 Jan 6;10(1):29-38. doi: 10.1016/j.celrep.2014.12.001. Epub 2014 Dec 24. Cell Rep. 2015. PMID: 25543141 Free PMC article.
-
Histone methylation and V(D)J recombination.
Shimazaki N, Lieber MR. Shimazaki N, et al. Int J Hematol. 2014 Sep;100(3):230-7. doi: 10.1007/s12185-014-1637-4. Epub 2014 Jul 25. Int J Hematol. 2014. PMID: 25060705 Review.
-
V(D)J recombination: RAG proteins, repair factors, and regulation.
Gellert M. Gellert M. Annu Rev Biochem. 2002;71:101-32. doi: 10.1146/annurev.biochem.71.090501.150203. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045092 Review.
Cited by
-
Daniel JA, Nussenzweig A. Daniel JA, et al. Biochim Biophys Acta. 2012 Jul;1819(7):733-8. doi: 10.1016/j.bbagrm.2012.01.019. Epub 2012 Feb 12. Biochim Biophys Acta. 2012. PMID: 22710321 Free PMC article. Review.
-
Maman Y, Teng G, Seth R, Kleinstein SH, Schatz DG. Maman Y, et al. Nucleic Acids Res. 2016 Nov 16;44(20):9624-9637. doi: 10.1093/nar/gkw633. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436288 Free PMC article.
-
The RAG2 C-terminus and ATM protect genome integrity by controlling antigen receptor gene cleavage.
Chaumeil J, Micsinai M, Ntziachristos P, Roth DB, Aifantis I, Kluger Y, Deriano L, Skok JA. Chaumeil J, et al. Nat Commun. 2013;4:2231. doi: 10.1038/ncomms3231. Nat Commun. 2013. PMID: 23900513 Free PMC article.
-
Unifying model for molecular determinants of the preselection Vβ repertoire.
Gopalakrishnan S, Majumder K, Predeus A, Huang Y, Koues OI, Verma-Gaur J, Loguercio S, Su AI, Feeney AJ, Artyomov MN, Oltz EM. Gopalakrishnan S, et al. Proc Natl Acad Sci U S A. 2013 Aug 20;110(34):E3206-15. doi: 10.1073/pnas.1304048110. Epub 2013 Aug 5. Proc Natl Acad Sci U S A. 2013. PMID: 23918392 Free PMC article.
-
Liu Y, Zhang L, Desiderio S. Liu Y, et al. Adv Exp Med Biol. 2009;650:157-65. doi: 10.1007/978-1-4419-0296-2_13. Adv Exp Med Biol. 2009. PMID: 19731809 Free PMC article. Review.
References
-
- Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases