Widespread microRNA repression by Myc contributes to tumorigenesis - PubMed
Widespread microRNA repression by Myc contributes to tumorigenesis
Tsung-Cheng Chang et al. Nat Genet. 2008 Jan.
Abstract
The c-Myc oncogenic transcription factor (Myc) is pathologically activated in many human malignancies. Myc is known to directly upregulate a pro-tumorigenic group of microRNAs (miRNAs) known as the miR-17-92 cluster. Through the analysis of human and mouse models of B cell lymphoma, we show here that Myc regulates a much broader set of miRNAs than previously anticipated. Unexpectedly, the predominant consequence of activation of Myc is widespread repression of miRNA expression. Chromatin immunoprecipitation reveals that much of this repression is likely to be a direct result of Myc binding to miRNA promoters. We further show that enforced expression of repressed miRNAs diminishes the tumorigenic potential of lymphoma cells. These results demonstrate that extensive reprogramming of the miRNA transcriptome by Myc contributes to tumorigenesis.
Figures

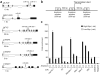
Repression of miRNA expression by Myc. (a) RNA blot analysis of miRNAs in P493-6 cells with high or low Myc expression. U6 small nuclear RNA (snRNA) was used as a loading control in this and all subsequent experiments (a representative blot is shown). ‘Expression ratio’ in this and subsequent figures indicates expression of the miRNA in the high-Myc state relative to the low-Myc state. ND, not detectable. (b) Organization of the human miR-30 clusters. miRNA clusters that are downregulated by Myc, as determined in c, are shown in bold. (c) RNA blots showing repression of miR-30 family members by Myc. Synthetic RNA oligonucleotides identical in sequence to each miR-30 family member and total RNA from P493-6 cells were hybridized with probes specific for each miRNA.
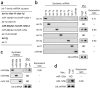
Myc associates with repressed pri-miRNA promoters. (a) Representations of repressed pri-miRNAs of known structure. (b) Real-time PCR amplicons for ChIP were designed within 250-bp windows immediately upstream of the transcription start site (amplicon S), 500-bp windows upstream of amplicon S (amplicon U) or 500-bp windows downstream of amplicon S (amplicon D). (c) Real-time PCR analysis of Myc chromatin immunoprecipitates. Fold enrichment in this and subsequent ChIP experiments represents the signal obtained after Myc immunoprecipitation relative to the signal obtained after immunoprecipitation with an irrelevant antibody. A validated Myc-bound amplicon in the promoter region of CDKN1A was used as a positive control. The 50-fold enrichment threshold for positive Myc binding is indicated as a broken line. Data are mean ± s.d. from three independent measurements.

Myc associates with conserved regions upstream of repressed miRNAs. (a) Phylogenetic conservation of the intergenic region containing the miR-29b-2/miR-29c cluster. Vista was used to generate pairwise alignments between the genomic sequence from human (May 2004 assembly) and that from the indicated species. The graph is a plot of nucleotide identity for a 100-bp sliding window centered at a given position. Annotated transcripts produced from this locus are shown at the top. Note that the 5′ end of miR-29b-2/miR-29c is toward the right. Arrows indicate locations of the real-time PCR amplicons used for ChIP experiments. (b) Real-time PCR analysis of Myc chromatin immunoprecipitates (see Fig. 2c). The conserved amplicon that showed maximal Myc binding (C) and a representative negative control amplicon (N) are shown for each miRNA. Locations of these and additional amplicons for the miR-29b-1/miR-29a cluster, the miR-30d/miR-30b cluster, miR-34a, miR-146a, the miR-195/miR-497 cluster and miR-150 are shown in Supplementary Figures 3-8. (c) Conserved Myc binding sites correspond to pri-miRNA promoters. Shown are the structures of pri-miRNA transcripts, as defined by 5′ and 3′ RACE. For some transcripts, alternative splicing gives rise to major and minor isoforms. Plots representing evolutionary conservation, shown below each transcript, were taken from the UCSC Genome Browser (human genome May 2004 assembly). Arrows indicate the locations of ChIP amplicons that yielded the highest Myc binding signals.
let-7 miRNAs are downregulated by Myc. (a) Organization of the human let-7 clusters. miRNA clusters downregulated by Myc, as determined in b-d, are shown in bold. (b-d) RNA blot analysis of synthetic RNA oligonucleotides or total RNA from P493-6 cells performed with probes specific for each member of the let-7 family (b), the miR-99/miR-100 family (c), and the miR-125 family (d). ND, not detectable.

Myc binds to conserved regions upstream of let-7 miRNAs. (a) Vista analysis of phylogenetic conservation encompassing the let-7a-1/let-7f-1/let-7d cluster, let-7g and the miR-99a/let-7c/miR-125b-2 cluster (see Fig. 3a). (b) Real-time PCR analysis of Myc chromatin immunoprecipitates (see Fig. 2c).
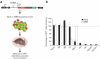
Expression of Myc-repressed miRNAs disadvantages lymphoma cell growth in vivo. (a) Myc3 or 38B9 lymphoma cells were infected with a retrovirus expressing a miRNA and GFP. The percentage of GFP-positive cells was measured before and after tumor formation. (b) Cells expressing select miRNAs are eliminated from tumors. Data are mean ± s.d. from three independent trials. At least 30% of cells were positive for GFP before injection into recipient mice.
Similar articles
-
Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation.
Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Chang TC, et al. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3384-9. doi: 10.1073/pnas.0808300106. Epub 2009 Feb 11. Proc Natl Acad Sci U S A. 2009. PMID: 19211792 Free PMC article.
-
Karkhanis V, Alinari L, Ozer HG, Chung J, Zhang X, Sif S, Baiocchi RA. Karkhanis V, et al. J Biol Chem. 2020 Jan 31;295(5):1165-1180. doi: 10.1074/jbc.RA119.008742. Epub 2019 Dec 10. J Biol Chem. 2020. PMID: 31822509 Free PMC article.
-
Targeting miR-21 with NL101 blocks c-Myc/Mxd1 loop and inhibits the growth of B cell lymphoma.
Li S, He X, Gan Y, Zhang J, Gao F, Lin L, Qiu X, Yu T, Zhang X, Chen P, Tong J, Qian W, Xu Y. Li S, et al. Theranostics. 2021 Jan 19;11(7):3439-3451. doi: 10.7150/thno.53561. eCollection 2021. Theranostics. 2021. PMID: 33537096 Free PMC article.
-
c-MYC-miRNA circuitry: a central regulator of aggressive B-cell malignancies.
Tao J, Zhao X, Tao J. Tao J, et al. Cell Cycle. 2014;13(2):191-8. doi: 10.4161/cc.27646. Epub 2014 Jan 6. Cell Cycle. 2014. PMID: 24394940 Free PMC article. Review.
-
Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs.
Robaina MC, Mazzoccoli L, Klumb CE. Robaina MC, et al. Cells. 2019 Oct 31;8(11):1365. doi: 10.3390/cells8111365. Cells. 2019. PMID: 31683676 Free PMC article. Review.
Cited by
-
Dang CV. Dang CV. Cell. 2012 Mar 30;149(1):22-35. doi: 10.1016/j.cell.2012.03.003. Cell. 2012. PMID: 22464321 Free PMC article. Review.
-
Circulating microRNA-144-5p is associated with depressive disorders.
Wang X, Sundquist K, Hedelius A, Palmér K, Memon AA, Sundquist J. Wang X, et al. Clin Epigenetics. 2015 Jul 22;7(1):69. doi: 10.1186/s13148-015-0099-8. eCollection 2015. Clin Epigenetics. 2015. PMID: 26199675 Free PMC article.
-
Xia B, Zhang L, Guo SQ, Li XW, Qu FL, Zhao HF, Zhang LY, Sun BC, You J, Zhang YZ. Xia B, et al. World J Gastroenterol. 2015 Feb 28;21(8):2433-42. doi: 10.3748/wjg.v21.i8.2433. World J Gastroenterol. 2015. PMID: 25741152 Free PMC article.
-
Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Wang Y, et al. J Biol Chem. 2013 Jun 21;288(25):18484-93. doi: 10.1074/jbc.M113.458158. Epub 2013 May 6. J Biol Chem. 2013. PMID: 23649629 Free PMC article.
-
The genetic architecture of multiple myeloma.
Morgan GJ, Walker BA, Davies FE. Morgan GJ, et al. Nat Rev Cancer. 2012 Apr 12;12(5):335-48. doi: 10.1038/nrc3257. Nat Rev Cancer. 2012. PMID: 22495321 Review.
References
-
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. - PubMed
-
- Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. - PubMed
-
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. - PubMed
-
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. - PubMed
-
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials