Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms - PubMed
- ️Tue Jan 01 2008
Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms
Xia Xu et al. Cancer Res. 2008.
Abstract
Ribonucleotide reductase (RNR) catalyzes the rate-limiting step in nucleotide biosynthesis and plays a central role in genome maintenance. Although a number of regulatory mechanisms govern RNR activity, the physiologic effect of RNR deregulation had not previously been examined in an animal model. We show here that overexpression of the small RNR subunit potently and selectively induces lung neoplasms in transgenic mice and is mutagenic in cultured cells. Combining RNR deregulation with defects in DNA mismatch repair, the cellular mutation correction system, synergistically increased RNR-induced mutagenesis and carcinogenesis. Moreover, the proto-oncogene K-ras was identified as a frequent mutational target in RNR-induced lung neoplasms. Together, these results show that RNR deregulation promotes lung carcinogenesis through a mutagenic mechanism and establish a new oncogenic activity for a key regulator of nucleotide metabolism. Importantly, RNR-induced lung neoplasms histopathologically resemble human papillary adenocarcinomas and arise stochastically via a mutagenic mechanism, making RNR transgenic mice a valuable model for lung cancer.
Figures
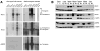
Northern blot analysis of RNR expression in wild-type and RNR transgenic mice. Total RNA was extracted from the indicated tissues from wild-type FVB mice (left panels), or RNR transgenic mice (right panels) and subjected to Northern blot hybridization with the indicated probes specific for Rrm1, Rrm2 or p53R2. Positions of endogenous and transgene-derived RNR transcripts are indicated. (B) Western blot analysis of RNR protein expression in the indicated tissues from wild type (WT) and RNR transgenic (Tg) mice, as well as lung neoplasms from the corresponding transgenic strains (Tumor 1, 2). Total protein from the indicated tissues was subjected to immunoblotting with antibodies specific to Rrm1, Rrm2 or p53R2. Duplicate membranes were immunoblotted for β-actin as a loading control.

(I) Lungs from a Rrm2Tg mouse with multiple independent neoplasms affecting several lobes. (II–VI) H&E-stained sections of lung neoplasms. (II) Solid adenoma from a p53R2Tg mouse. (III–VI) Papillary adenocarcinomas from Rrm2Tg or p53R2Tg mice showing pleural invasion (arrow) (III), regional variation in growth pattern (IV), multiple mitotic figures (arrows) (V), and blood vessel invasion (arrow) (VI). (VII, VIII) Immunohistochemical staining of RNR-induced lung neoplasms for Pro-SP-C (VII) or CC10 (VIII) by the ABC method, with methyl green counterstain. Inserts show higher magnification views of the boxed regions. Calibration bar: II, IV: 50 μm; III: 241 μm; V: 10 μm; VI: 25 μm; VII, VIII: 100 μm. (B) Northern blot analysis of lung neoplasms from RNR transgenic mice. Total RNA was prepared from lung neoplasms (Tumor 1, Tumor 2, Tumor 3) or normal lung tissue (Lung) from RNR transgenic mice, as well as from wild-type FVB lung tissue (WT FVB). Northern blotting was performed with the indicated radiolabeled probes.
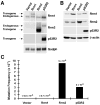
Northern blot analysis of RNR expression in stable 3T3 cell pools transfected with either pCaggs empty vector or pCaggs RNR genes. Total RNA was extracted from the indicated cell lines and subjected to Northern blot hybridization with probes specific for Rrm1, Rrm2, p53R2, or Gapdh. (B) Western blot analysis of RNR protein expression in RNR overexpressing 3T3 cells. Total protein was extracted from the indicated cell lines and subjected to immunoblotting with antibodies specific to Rrm1, Rrm2, or p53R2. Duplicate membranes were immunoblotted for β-actin as a loading control. Samples in (A) and (B) were run on single blots, which were then cropped to remove extraneous lanes. (C) Mutation frequency at the Hprt locus in Rrm1, Rrm2 and p53R2 overexpressing 3T3 cells. Mutation frequency was determined by Hprt assay.

Canavanine mutation rate assay for RNR1(WT) and rnr1-D57N strains on MMR-deficient backgrounds (msh3Δ, msh2Δ, msh6Δ, or WT) of S. cerevisiae. The forward mutation rate (per generation) to canavanine resistance was measured for the indicated single and double mutant combinations. Error bars show the 95% confidence interval. (B) Survival curves for Msh6−/−RNRTg (Rrm2Tg or p53R2Tg) mice. Mice were aged until moribund for up to 17 months. Survival curves were generated using SPSS software. The following number of animals was analyzed for each genotype: Msh+/+ (11), Msh6+/− (23), Msh6+/−Rrm2Tg (22), Msh6+/−p53R2Tg (11), Msh6−/− (34), Msh6−/−Rrm2Tg (20), Msh6−/− p53R2Tg (17). (C, D) Mutation frequency at the λ cII locus in lung (C) or spleen (D) tissues from RNR overexpressing and control mice. Genomic DNA was isolated from 3-month old mice of the indicated genotypes and packaged into infectious phage. Mutation frequency was determined based on the ratio of the number of mutant phage obtained to the total number of phage analyzed.
Similar articles
-
Ribonucleotide reductase is not limiting for mitochondrial DNA copy number in mice.
Ylikallio E, Page JL, Xu X, Lampinen M, Bepler G, Ide T, Tyynismaa H, Weiss RS, Suomalainen A. Ylikallio E, et al. Nucleic Acids Res. 2010 Dec;38(22):8208-18. doi: 10.1093/nar/gkq735. Epub 2010 Aug 19. Nucleic Acids Res. 2010. PMID: 20724444 Free PMC article.
-
Role of Ribonucleotide Reductase in Bacillus subtilis Stress-Associated Mutagenesis.
Castro-Cerritos KV, Yasbin RE, Robleto EA, Pedraza-Reyes M. Castro-Cerritos KV, et al. J Bacteriol. 2017 Jan 30;199(4):e00715-16. doi: 10.1128/JB.00715-16. Print 2017 Feb 15. J Bacteriol. 2017. PMID: 27920297 Free PMC article.
-
Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents.
Cerqueira NM, Fernandes PA, Ramos MJ. Cerqueira NM, et al. Recent Pat Anticancer Drug Discov. 2007 Jan;2(1):11-29. doi: 10.2174/157489207779561408. Recent Pat Anticancer Drug Discov. 2007. PMID: 18221051 Review.
-
DNA damage and cell cycle regulation of ribonucleotide reductase.
Elledge SJ, Zhou Z, Allen JB, Navas TA. Elledge SJ, et al. Bioessays. 1993 May;15(5):333-9. doi: 10.1002/bies.950150507. Bioessays. 1993. PMID: 8343143 Review.
Cited by
-
Tang X, Xu Y, Lu L, Jiao Y, Liu J, Wang L, Zhao H. Tang X, et al. Cancer Manag Res. 2018 Sep 13;10:3533-3549. doi: 10.2147/CMAR.S171661. eCollection 2018. Cancer Manag Res. 2018. PMID: 30271202 Free PMC article.
-
D'Angiolella V, Esencay M, Pagano M. D'Angiolella V, et al. Trends Cell Biol. 2013 Mar;23(3):135-40. doi: 10.1016/j.tcb.2012.10.011. Epub 2012 Nov 19. Trends Cell Biol. 2013. PMID: 23182110 Free PMC article. Review.
-
Ribonucleotide reductase is not limiting for mitochondrial DNA copy number in mice.
Ylikallio E, Page JL, Xu X, Lampinen M, Bepler G, Ide T, Tyynismaa H, Weiss RS, Suomalainen A. Ylikallio E, et al. Nucleic Acids Res. 2010 Dec;38(22):8208-18. doi: 10.1093/nar/gkq735. Epub 2010 Aug 19. Nucleic Acids Res. 2010. PMID: 20724444 Free PMC article.
-
Inhibitors of the Cancer Target Ribonucleotide Reductase, Past and Present.
Huff SE, Winter JM, Dealwis CG. Huff SE, et al. Biomolecules. 2022 Jun 10;12(6):815. doi: 10.3390/biom12060815. Biomolecules. 2022. PMID: 35740940 Free PMC article. Review.
-
Wang Q, Liu X, Zhou J, Huang Y, Zhang S, Shen J, Loera S, Yuan X, Chen W, Jin M, Shibata S, Liu Y, Chu P, Wang L, Yen Y. Wang Q, et al. PLoS One. 2013 Jul 29;8(7):e70191. doi: 10.1371/journal.pone.0070191. Print 2013. PLoS One. 2013. PMID: 23922955 Free PMC article.
References
-
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. - PubMed
-
- Tanaka H, Arakawa H, Yamaguchi T, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–9. - PubMed
-
- Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–9. - PubMed
-
- Thelander L. Ribonucleotide reductase and mitochondrial DNA synthesis. Nat Genet. 2007;39:703–4. - PubMed
-
- Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM053085-13/GM/NIGMS NIH HHS/United States
- T32GM07617/GM/NIGMS NIH HHS/United States
- T32 GM007273-33/GM/NIGMS NIH HHS/United States
- R56 CA096823/CA/NCI NIH HHS/United States
- R01 CA096823/CA/NCI NIH HHS/United States
- CA96823/CA/NCI NIH HHS/United States
- R01 CA108773-04/CA/NCI NIH HHS/United States
- R01 CA096823-05/CA/NCI NIH HHS/United States
- K26 RR017595/RR/NCRR NIH HHS/United States
- T32 GM007617/GM/NIGMS NIH HHS/United States
- K26 RR017595-05/RR/NCRR NIH HHS/United States
- R01 GM053085/GM/NIGMS NIH HHS/United States
- GM53085/GM/NIGMS NIH HHS/United States
- T32 GM007273/GM/NIGMS NIH HHS/United States
- R01 CA108773/CA/NCI NIH HHS/United States
- RR017595/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous