Fast, accurate and simulation-free stochastic mapping - PubMed
- ️Tue Jan 01 2008
Fast, accurate and simulation-free stochastic mapping
Vladimir N Minin et al. Philos Trans R Soc Lond B Biol Sci. 2008.
Abstract
Mapping evolutionary trajectories of discrete traits onto phylogenies receives considerable attention in evolutionary biology. Given the trait observations at the tips of a phylogenetic tree, researchers are often interested where on the tree the trait changes its state and whether some changes are preferential in certain parts of the tree. In a model-based phylogenetic framework, such questions translate into characterizing probabilistic properties of evolutionary trajectories. Current methods of assessing these properties rely on computationally expensive simulations. In this paper, we present an efficient, simulation-free algorithm for computing two important and ubiquitous evolutionary trajectory properties. The first is the mean number of trait changes, where changes can be divided into classes of interest (e.g. synonymous/non-synonymous mutations). The mean evolutionary reward, accrued proportionally to the time a trait occupies each of its states, is the second property. To illustrate the usefulness of our results, we first employ our simulation-free stochastic mapping to execute a posterior predictive test of correlation between two evolutionary traits. We conclude by mapping synonymous and non-synonymous mutations onto branches of an HIV intrahost phylogenetic tree and comparing selection pressure on terminal and internal tree branches.
Figures
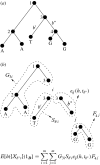
Sandwich formula illustration. (a) An example phylogenetic tree in which we label internal nodes numerically and two branches b* and b′. We break this tree at nodes 3 and 4 into the subtrees shown in (b). Assuming that the trait states are i and j at nodes 3 and 4, respectively, we mark each subtree by the corresponding quantity needed for calculating the posterior expectation of a mapping summary on branch b*.

Primate trait data. We plot a phylogenetic tree, randomly chosen from the posterior sample, of 60 primate species. Branches of the sampled tree are not drawn to scale, nor is the tree ultrametric. Taxa names and trait values (‘0’, absence; ‘1’, presence; ‘−’, missing) for oestrus advertisement (EA) and multimale mating system (MS) are depicted at the tips of the tree.
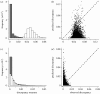
Testing coevolution. (a,c) Plots depicting observed (white bars) and predicted (grey bars) distributions of the discrepancy measure for the (a,b) primate and (c,d) simulated data. (b,d) The scatter plots of these distributions.
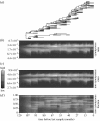
Time evolution of synonymous and non-synonymous rates. (a) A representative phylogeny of 129 intrahost HIV sequences. The three heat maps depict the marginal posterior densities of the (b) synonymous and (c) non-synonymous rates, and (d) the proportion of non-synonymous mutations over time.
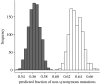
Bimodality of the fraction of non-synonymous mutations. We plot the predicted fraction of non-synonymous mutations computed for terminal (grey bars) and internal (white bars) branches.
Similar articles
-
Calculating Higher-Order Moments of Phylogenetic Stochastic Mapping Summaries in Linear Time.
Dhar A, Minin VN. Dhar A, et al. J Comput Biol. 2017 May;24(5):377-399. doi: 10.1089/cmb.2016.0172. Epub 2017 Feb 8. J Comput Biol. 2017. PMID: 28177780 Free PMC article.
-
SFREEMAP - A simulation-free tool for stochastic mapping.
Pasqualin D, Barbeitos M, Silva F. Pasqualin D, et al. BMC Bioinformatics. 2017 Feb 22;18(1):123. doi: 10.1186/s12859-017-1554-7. BMC Bioinformatics. 2017. PMID: 28228094 Free PMC article.
-
A Cooperative Co-Evolutionary Genetic Algorithm for Tree Scoring and Ancestral Genome Inference.
Gao N, Zhang Y, Feng B, Tang J. Gao N, et al. IEEE/ACM Trans Comput Biol Bioinform. 2015 Nov-Dec;12(6):1248-54. doi: 10.1109/TCBB.2015.2430860. IEEE/ACM Trans Comput Biol Bioinform. 2015. PMID: 26671797
-
Efficient selection of branch-specific models of sequence evolution.
Dutheil JY, Galtier N, Romiguier J, Douzery EJ, Ranwez V, Boussau B. Dutheil JY, et al. Mol Biol Evol. 2012 Jul;29(7):1861-74. doi: 10.1093/molbev/mss059. Epub 2012 Feb 2. Mol Biol Evol. 2012. PMID: 22319139
-
Bayesian coestimation of phylogeny and sequence alignment.
Lunter G, Miklós I, Drummond A, Jensen JL, Hein J. Lunter G, et al. BMC Bioinformatics. 2005 Apr 1;6:83. doi: 10.1186/1471-2105-6-83. BMC Bioinformatics. 2005. PMID: 15804354 Free PMC article.
Cited by
-
Jacquot M, Wallace MA, Streicker DG, Biek R. Jacquot M, et al. Viruses. 2022 Oct 29;14(11):2399. doi: 10.3390/v14112399. Viruses. 2022. PMID: 36366496 Free PMC article.
-
Untangling lineage introductions, persistence and transmission drivers of HP-PRRSV sublineage 8.7.
Sun Y, Xing J, Hong SL, Bollen N, Xu S, Li Y, Zhong J, Gao X, Zhu D, Liu J, Gong L, Zhou L, An T, Shi M, Wang H, Baele G, Zhang G. Sun Y, et al. Nat Commun. 2024 Oct 13;15(1):8842. doi: 10.1038/s41467-024-53076-w. Nat Commun. 2024. PMID: 39397015 Free PMC article.
-
Pérez AB, Vrancken B, Chueca N, Aguilera A, Reina G, García-Del Toro M, Vera F, Von Wichman MA, Arenas JI, Téllez F, Pineda JA, Omar M, Bernal E, Rivero-Juárez A, Fernández-Fuertes E, de la Iglesia A, Pascasio JM, Lemey P, Garcia F, Cuypers L. Pérez AB, et al. Euro Surveill. 2019 Feb;24(9):1800227. doi: 10.2807/1560-7917.ES.2019.24.9.1800227. Euro Surveill. 2019. PMID: 30862327 Free PMC article.
-
Emerging Concepts of Data Integration in Pathogen Phylodynamics.
Baele G, Suchard MA, Rambaut A, Lemey P. Baele G, et al. Syst Biol. 2017 Jan 1;66(1):e47-e65. doi: 10.1093/sysbio/syw054. Syst Biol. 2017. PMID: 28173504 Free PMC article. Review.
-
Lemey P, Ruktanonchai N, Hong SL, Colizza V, Poletto C, Van den Broeck F, Gill MS, Ji X, Levasseur A, Sadilek A, Lai S, Tatem AJ, Baele G, Suchard MA, Dellicour S. Lemey P, et al. Res Sq [Preprint]. 2021 Feb 10:rs.3.rs-208849. doi: 10.21203/rs.3.rs-208849/v1. Res Sq. 2021. PMID: 33594355 Free PMC article. Updated. Preprint.
References
-
- Ball F, Milne R. Simple derivations of properties of counting processes associated with Markov renewal processes. J. Appl. Prob. 2005;42:1031–1043. doi:10.1239/jap/1134587814 - DOI
-
- Cannings, C., Thompson, E. & Skolnick, M. 1980 Pedigree analysis of complex models. In Current developments in anthropological genetics, pp. 251–298. New York, NY: Plenum Press.
-
- Dimmic M, Hubisz M, Bustamante C, Nielsen R. Detecting coevolving amino acid sites using Bayesian mutational mapping. Bioinformatics. 2005;21:i126–i135. doi:10.1093/bioinformatics/bti1032 - DOI - PubMed
-
- Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi:10.1186/1471-2148-7-214 - DOI - PMC - PubMed
-
- Dutheil J, Pupko T, Marie A, Galtier N. A model-based approach for detecting coevolving positions in a molecule. Mol. Biol. Evol. 2005;22:1919–1928. doi:10.1093/molbev/msi183 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources