Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network - PubMed
. 2009 Dec;5(12):e1000777.
doi: 10.1371/journal.pgen.1000777. Epub 2009 Dec 18.
Tali Mandel, Shira Mintz-Oron, Ilya Venger, Dorit Levy, Merav Yativ, Eva Domínguez, Zhonghua Wang, Ric C H De Vos, Reinhard Jetter, Lukas Schreiber, Antonio Heredia, Ilana Rogachev, Asaph Aharoni
Affiliations
- PMID: 20019811
- PMCID: PMC2788616
- DOI: 10.1371/journal.pgen.1000777
Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network
Avital Adato et al. PLoS Genet. 2009 Dec.
Abstract
The cuticle covering plants' aerial surfaces is a unique structure that plays a key role in organ development and protection against diverse stress conditions. A detailed analysis of the tomato colorless-peel y mutant was carried out in the framework of studying the outer surface of reproductive organs. The y mutant peel lacks the yellow flavonoid pigment naringenin chalcone, which has been suggested to influence the characteristics and function of the cuticular layer. Large-scale metabolic and transcript profiling revealed broad effects on both primary and secondary metabolism, related mostly to the biosynthesis of phenylpropanoids, particularly flavonoids. These were not restricted to the fruit or to a specific stage of its development and indicated that the y mutant phenotype is due to a mutation in a regulatory gene. Indeed, expression analyses specified three R2R3-MYB-type transcription factors that were significantly down-regulated in the y mutant fruit peel. One of these, SlMYB12, was mapped to the genomic region on tomato chromosome 1 previously shown to harbor the y mutation. Identification of an additional mutant allele that co-segregates with the colorless-peel trait, specific down-regulation of SlMYB12 and rescue of the y phenotype by overexpression of SlMYB12 on the mutant background, confirmed that a lesion in this regulator underlies the y phenotype. Hence, this work provides novel insight to the study of fleshy fruit cuticular structure and paves the way for the elucidation of the regulatory network that controls flavonoid accumulation in tomato fruit cuticle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
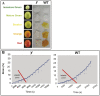
(A) Enzymatically isolated cuticles (n = 20) of wt and y mutant at five stages of fruit development. (B) Biomechanical tests of isolated fruit cuticles (n = 7) at the red stage of development reveal significant differences in the elastic phase between wt and y mutant cuticles.
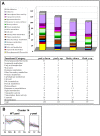
(A) Functional category distribution among differentially expressed wt and y mutant transcripts at the three latest stages of fruit development. (B) The expression profile (obtained by array analysis) of genes belonging to cluster 14 (total 38 members) in the peel tissue of y mutant and wt fruit. In the y mutant, array analysis was carried out on three of the five stages of fruit development examined in the wt fruit.

(A) PCA of metabolic profiles obtained by GC-MS analysis, with samples of wt and y peel and flesh tissues at five stages of fruit development (n = 3). (B) PCA of metabolic profiles obtained by UPLC-QTOF-MS analysis, with samples of wt and y peel and flesh tissues in the last three stages of fruit development (n = 3). Distinct metabolic profiles that correspond to particular stages of y and wt fruit development are encircled in (A,B).

Indicated by asterisks are significant differences as analyzed by two-way ANOVA and post-hoc analyses, see Materials and Methods (n = 3 for each sample). Y axis indicates metabolites' relative quantification by normalization of their response values to the Ribitol internal standard (IS) (see also Mintz-Oron et al. [6]).
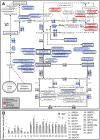
(A) Changes in gene expression (detected using array and/or real-time PCR analyses) and metabolite levels (detected by UPLC-QTOF-MS and GC-MS analyses) in tomato fruit peel. Red and blue colors represent up- and down-regulation, respectively. (B) Real-time PCR relative expression analyses of selected transcripts from the phenylpropanoid pathway in wt and y mutant tomato peels at the breaker stage of fruit development. Indicated by asterisks are significant differences analyzed by Student's t-test (n = 3; P<0.05; bars indicate standard errors). Gene identifiers and RT–PCR primers are listed in Table S3. #, caffeoylquinic acid derivatives that include: 4-caffeoylquinic acid, 5-caffeoylquinic acid, dicaffeoylquinic acid I/II/III, tricaffeoylquinic acid and 3-dicaffeoylquinic acid, detected but not altered in both peel and flesh of the y mutant. Metabolites marked by * or ** may be NarCh or Nar derivatives.
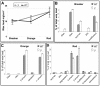
(A) Total cuticular wax load expressed as mg/cm2. (B) Amounts (%) relative to total wax coverage of individual compounds that were significantly altered at the breaker stage of fruit development, (C) at the orange stage of fruit development, and (D) at the ripe red stage of fruit development. 29an, C29 alkane; 30an, C30 alkane; iso31an, C31 iso alkane; 24ic, tetracosanoic acid.

(A) HPLC-PDA analysis detected six isoprenoids that differ significantly between the wt and y mutant fruit tissues at the orange stage of fruit development. Indicated by asterisks are significant differences analyzed by Student's t-test (n = 3; P<0.05; bars indicate standard errors). (B) Changes in isoprenoid-related gene expression (detected by array analysis) and metabolite levels (detected by HPLC-PDA) in tomato fruit peel and flesh tissues. Red and blue colors represent up- and down-regulation, respectively. Significant down-regulation of SlCRTR-B2 and SlNCED transcript levels in the y mutant peel was confirmed by RT–PCR analyses (Student's t-test, n = 3; P<0.05; bars indicate standard errors). Primers, transcript identifiers/accessions and expression data are listed in Table S4.
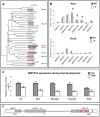
(A) Phylogenetic analysis of the putative tomato regulators reported in our study and known phenylpropanoid/flavonoid-related transcription factors from other species. The ClustalX and NJplot softwares were used to compute the tree and its significance (bootstrap) values. (B) RT–PCR relative expression analyses of selected tomato transcription factors putatively related to the regulation of the phenylpropanoid/flavonoid pathway in wt and y mutant tissues at the breaker stage of fruit development. Indicated by asterisks are significant differences analyzed by Student's t-test (n = 3; P<0.05; bars indicate standard errors). Gene identifiers and primers are listed in Table S3. (C) Real Time RT–PCR relative expression analysis of SlMYB12 in wt fruit tissues through five developmental stages reveals a peel-associated expression pattern. IG, Immature Green; MG, Mature Green; Br, Breaker; Or, Orange; Re, Red, stages of fruit development (n = 3; P<0.05; bars indicate standard errors). (D) Structure of the SlMYB12 gene. Gray regions represent coding sequence and white represents UTRs. Arrows indicate the position of RT–PCR primers, red brackets indicate the positions of premature polyadenylation sites (in the y-1 mutant allele), and the black bracket indicates the position of the alternative wt polyadenylation site.
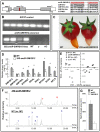
(A) Red box indicates the location of the amiR-SlMYB12 target sequence on the SlMYB12 gene, arrows indicate the position of RT–PCR primers, and the sequence alignment on the right demonstrates the specificity of this artificial microRNA. (B) Expression of the amiR-SlMYB12 precursor in samples extracted from leaves of 35S:amiR-SlMYB12-transgenic lines and non-transgenic controls. (C) Fruit of 35S:amiR-SlMYB12-transgenic lines display colorless peel. (D) RT–PCR relative expression analysis of phenylpropanoid/flavonoid-related regulators and structural genes in fruit peel of wt and 35S:amiR-SlMYB12-transgenic lines. Indicated by asterisks are significantly reduced levels analyzed by Student's t-test (n = 3; P<0.05; bars indicate standard errors). (E) PCA of metabolic profiles obtained by UPLC-QTOF-MS analysis carried out on peel samples of wt cv. AC and cv. MT, y mutant and an amiR-SlMYB12-transgenic line at the red stage of fruit development. Analysis was performed with the TMEV program using normalized and log-transformed data. (F) Total ion chromatograms (TICs) of wt (cv. MT) and 35S:amiR-SlMYB12 peels at the red stage of fruit development, acquired in the negative mode using UPLC-QTOF-MS (in relative intensity, 100% corresponds to 6.14×104 counts). The putative identity of the differential compounds is: 1 - quercetin-dihexose-deoxyhexose, 2 - quercetin-hexose-deoxyhexose-pentose, 3 - quercetin-rutinoside (rutin), 4 - phloretin-di-C-hexose, 5 - kaempferol-glucose-rhamnose, 6 - naringenin chalcone, 7 - dicaffeoylquinic acid III, 8 - tricaffeoylquinic acid. Red and blue numbers indicate metabolites that showed elevated or reduced levels in the transgene samples in comparison to those of their corresponding wt, respectively. (G) Relative levels of NarCh in cv. MT and 35S:amiR-SlMYB12, expressed as chromatographic peak areas, calculated for m/z 271.06 Da (n = 5).
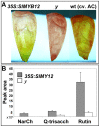
(A) Peels of red fruit from the 35S:SlMYB12 line on a y background, the y mutant and wt cv. AC. (B) UPLC-PDA analysis of red fruit peels reveals significantly different levels of flavanoids between regions of phenotype complementation in peels of the 35S:SlMYB12 line and those of the y mutant (n = 3; P<0.01; bars represent standard error). The UPLC instrument used in this analysis is equipped with an Acquity 2996 PDA detector. Sample preparation and LC conditions were as described for the UPLC-QTOF-MS analysis. Compounds peak areas were determined by Empower 2 software (Waters) at 370 nm for naringenin chalcone (NarCh) and at 256 nm for quercetin-hexose-deoxyhexose-pentose (Q-trisacch) and quercetin-rutinoside (rutin).
Similar articles
-
Ballester AR, Molthoff J, de Vos R, Hekkert Bt, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, Ykema M, Granell A, Bovy A. Ballester AR, et al. Plant Physiol. 2010 Jan;152(1):71-84. doi: 10.1104/pp.109.147322. Epub 2009 Nov 11. Plant Physiol. 2010. PMID: 19906891 Free PMC article.
-
Fernandez-Moreno JP, Tzfadia O, Forment J, Presa S, Rogachev I, Meir S, Orzaez D, Aharoni A, Granell A. Fernandez-Moreno JP, et al. Plant Physiol. 2016 Jul;171(3):1821-36. doi: 10.1104/pp.16.00282. Epub 2016 May 6. Plant Physiol. 2016. PMID: 27208285 Free PMC article.
-
Gene expression and metabolism in tomato fruit surface tissues.
Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, Aharoni A. Mintz-Oron S, et al. Plant Physiol. 2008 Jun;147(2):823-51. doi: 10.1104/pp.108.116004. Epub 2008 Apr 25. Plant Physiol. 2008. PMID: 18441227 Free PMC article.
-
Chattopadhyay T, Hazra P, Akhtar S, Maurya D, Mukherjee A, Roy S. Chattopadhyay T, et al. Plant Cell Rep. 2021 May;40(5):767-782. doi: 10.1007/s00299-020-02650-9. Epub 2021 Jan 3. Plant Cell Rep. 2021. PMID: 33388894 Review.
Cited by
-
Xie Q, Liu Z, Meir S, Rogachev I, Aharoni A, Klee HJ, Galili G. Xie Q, et al. Plant Biotechnol J. 2016 Dec;14(12):2300-2309. doi: 10.1111/pbi.12583. Epub 2016 Jun 22. Plant Biotechnol J. 2016. PMID: 27185473 Free PMC article.
-
Tomato Fruit Development and Metabolism.
Quinet M, Angosto T, Yuste-Lisbona FJ, Blanchard-Gros R, Bigot S, Martinez JP, Lutts S. Quinet M, et al. Front Plant Sci. 2019 Nov 29;10:1554. doi: 10.3389/fpls.2019.01554. eCollection 2019. Front Plant Sci. 2019. PMID: 31850035 Free PMC article. Review.
-
Vilperte V, Lucaciu CR, Halbwirth H, Boehm R, Rattei T, Debener T. Vilperte V, et al. BMC Genomics. 2019 Nov 27;20(1):900. doi: 10.1186/s12864-019-6247-3. BMC Genomics. 2019. PMID: 31775622 Free PMC article.
-
Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, Fernie AR, Fei Z, Giovannoni JJ, Rose JK. Matas AJ, et al. Plant Cell. 2011 Nov;23(11):3893-910. doi: 10.1105/tpc.111.091173. Epub 2011 Nov 1. Plant Cell. 2011. PMID: 22045915 Free PMC article.
-
Engl C, Waite CJ, McKenna JF, Bennett MH, Hamann T, Buck M. Engl C, et al. mBio. 2014 May 20;5(3):e01168-14. doi: 10.1128/mBio.01168-14. mBio. 2014. PMID: 24846383 Free PMC article.
References
-
- Samuels L, Jetter R, Kunst L. First steps in understanding the export of lipids to the plant cuticle. Plant Biosystems. 2005;139:65–68.
-
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nature Biotechnology. 2001;19:470–474. - PubMed
-
- Luque P, Bruque S, Heredia A. Water permeability of isolated cuticular membranes: a structural analysis. Arch Biochem Biophys. 1995;317:417–422. - PubMed
-
- Willits MG, Kramer CM, Prata RT, De Luca V, Potter BG, et al. Utilization of the genetic resources of wild species to create a nontransgenic high flavonoid tomato. J Agric Food Chem. 2005;53:1231–1236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases