PETcofold: predicting conserved interactions and structures of two multiple alignments of RNA sequences - PubMed
- ️Sat Jan 01 2011
PETcofold: predicting conserved interactions and structures of two multiple alignments of RNA sequences
Stefan E Seemann et al. Bioinformatics. 2011.
Abstract
Motivation: Predicting RNA-RNA interactions is essential for determining the function of putative non-coding RNAs. Existing methods for the prediction of interactions are all based on single sequences. Since comparative methods have already been useful in RNA structure determination, we assume that conserved RNA-RNA interactions also imply conserved function. Of these, we further assume that a non-negligible amount of the existing RNA-RNA interactions have also acquired compensating base changes throughout evolution. We implement a method, PETcofold, that can take covariance information in intra-molecular and inter-molecular base pairs into account to predict interactions and secondary structures of two multiple alignments of RNA sequences.
Results: PETcofold's ability to predict RNA-RNA interactions was evaluated on a carefully curated dataset of 32 bacterial small RNAs and their targets, which was manually extracted from the literature. For evaluation of both RNA-RNA interaction and structure prediction, we were able to extract only a few high-quality examples: one vertebrate small nucleolar RNA and four bacterial small RNAs. For these we show that the prediction can be improved by our comparative approach. Furthermore, PETcofold was evaluated on controlled data with phylogenetically simulated sequences enriched for covariance patterns at the interaction sites. We observed increased performance with increased amounts of covariance.
Availability: The program PETcofold is available as source code and can be downloaded from http://rth.dk/resources/petcofold.
Figures
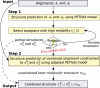
The
PETcofoldpipeline consists of two steps: (1) intra-molecular folding by
PETfoldand selection of a set of highly reliable base pairs that only decreases the probability of the ensemble in some pre-defined range; (2) inter-molecular folding by adapted
PETfoldusing constraints from step 1. In the end, partial structures and constrained inter-molecular structures are combined to form the joint RNA secondary structure including pseudoknots.
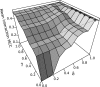
The performance of
PETcofoldwhile varying the parameters δ (maximal intra-molecular base pair reliability) and γ (minimal partial structure probability). The 3D plot shows the mean MCC of 32 interactions using input sequences with a minimal pairwise mRNA interaction site sequence identity to the reference of 60%. Predictions were carried out without the option -noLP. The maximal MCC is marked with ‘+’.
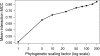
Prediction performance of
PETcofoldon phylogenetically simulated sequence data for 32 interactions. Covariance of the data were increased by multiplying each branch length with a phylogenetic scaling factor.
PETcofoldwas called with parameters δ = 0.9, γ = 0.1 and option -noLP.
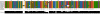
Joint secondary structure of the sRNA MicA and the mRNA ompA. The alignment shows the two input alignments concatenated by the linker symbol ‘&’, the joint structure predicted by
PETcofold(with parameters δ = 0.9, γ = 0.1 and option -noLP) and the model of the MicA–ompA complex proposed by Udekwu et al. (2005). Sequences are labelled with the genome accession numbers of the corresponding organisms. Angle brackets indicate inter-molecular base pairs. Round and square brackets indicate intra-molecular base pairs. Square brackets indicate positions that are constrained in step 1 of the
PETcofoldpipeline. For ompA, only 40 nt upstream of the interaction site are shown. The alignment was visualized with
Jalview(Waterhouse et al., 2009).
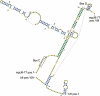
Joint secondary structure of mgU6-77 snoRNA and U6 snRNA as predicted by
PETcofoldusing parameters δ = 0.9, γ = 0.1 and options -noLP and -extstem. Mouse sequences are shown, but the numbering refers to the input alignment of human, chimp, mouse, cow, tenrec, dog and opossum. Intra-molecular base pairs are coloured in blue and inter-molecular base pairs in green. The cytosine at alignment position 78 (pos. 77 in mouse) is 2′-O-methylated and marked by ‘m’. We used
PseudoViewer3for drawing (Byun and Han, 2009).
Similar articles
-
Seemann SE, Menzel P, Backofen R, Gorodkin J. Seemann SE, et al. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W107-11. doi: 10.1093/nar/gkr248. Epub 2011 May 23. Nucleic Acids Res. 2011. PMID: 21609960 Free PMC article.
-
RILogo: visualizing RNA-RNA interactions.
Menzel P, Seemann SE, Gorodkin J. Menzel P, et al. Bioinformatics. 2012 Oct 1;28(19):2523-6. doi: 10.1093/bioinformatics/bts461. Epub 2012 Jul 23. Bioinformatics. 2012. PMID: 22826541
-
MASTR: multiple alignment and structure prediction of non-coding RNAs using simulated annealing.
Lindgreen S, Gardner PP, Krogh A. Lindgreen S, et al. Bioinformatics. 2007 Dec 15;23(24):3304-11. doi: 10.1093/bioinformatics/btm525. Epub 2007 Nov 15. Bioinformatics. 2007. PMID: 18006551
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.
Washietl S, Bernhart SH, Kellis M. Washietl S, et al. Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
Sequence and structure analysis of noncoding RNAs.
Washietl S. Washietl S. Methods Mol Biol. 2010;609:285-306. doi: 10.1007/978-1-60327-241-4_17. Methods Mol Biol. 2010. PMID: 20221926 Review.
Cited by
-
Direct updating of an RNA base-pairing probability matrix with marginal probability constraints.
Hamada M. Hamada M. J Comput Biol. 2012 Dec;19(12):1265-76. doi: 10.1089/cmb.2012.0215. J Comput Biol. 2012. PMID: 23210474 Free PMC article.
-
Conserved RNA secondary structures and long-range interactions in hepatitis C viruses.
Fricke M, Dünnes N, Zayas M, Bartenschlager R, Niepmann M, Marz M. Fricke M, et al. RNA. 2015 Jul;21(7):1219-32. doi: 10.1261/rna.049338.114. Epub 2015 May 11. RNA. 2015. PMID: 25964384 Free PMC article.
-
Seemann SE, Menzel P, Backofen R, Gorodkin J. Seemann SE, et al. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W107-11. doi: 10.1093/nar/gkr248. Epub 2011 May 23. Nucleic Acids Res. 2011. PMID: 21609960 Free PMC article.
-
How to do RNA-RNA Interaction Prediction? A Use-Case Driven Handbook Using IntaRNA.
Raden M, Miladi M. Raden M, et al. Methods Mol Biol. 2024;2726:209-234. doi: 10.1007/978-1-0716-3519-3_9. Methods Mol Biol. 2024. PMID: 38780733 Review.
-
IRBIS: a systematic search for conserved complementarity.
Pervouchine DD. Pervouchine DD. RNA. 2014 Oct;20(10):1519-31. doi: 10.1261/rna.045088.114. Epub 2014 Aug 20. RNA. 2014. PMID: 25142064 Free PMC article.
References
-
- Alkan C, et al. RNA-RNA interaction prediction and antisense RNA target search. J. Comput. Biol. 2006;13:267–282. - PubMed
-
- Andronescu M, et al. Secondary structure prediction of interacting RNA molecules. J. Mol. Biol. 2005;345:987–1001. - PubMed
-
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. - PubMed
-
- Bachellerie JP, et al. The expanding snoRNA world. Biochimie. 2002;84:775–790. - PubMed