The effectiveness of three regions in mitochondrial genome for aphid DNA barcoding: a case in Lachininae - PubMed
The effectiveness of three regions in mitochondrial genome for aphid DNA barcoding: a case in Lachininae
Rui Chen et al. PLoS One. 2012.
Erratum in
- PLoS One. 2013;8(5). doi:10.1371/annotation/3595dc99-a0fc-47fc-b21b-b54003054ffd
Abstract
Background: The mitochondrial gene COI has been widely used by taxonomists as a standard DNA barcode sequence for the identification of many animal species. However, the COI region is of limited use for identifying certain species and is not efficiently amplified by PCR in all animal taxa. To evaluate the utility of COI as a DNA barcode and to identify other barcode genes, we chose the aphid subfamily Lachninae (Hemiptera: Aphididae) as the focus of our study. We compared the results obtained using COI with two other mitochondrial genes, COII and Cytb. In addition, we propose a new method to improve the efficiency of species identification using DNA barcoding.
Methodology/principal findings: Three mitochondrial genes (COI, COII and Cytb) were sequenced and were used in the identification of over 80 species of Lachninae. The COI and COII genes demonstrated a greater PCR amplification efficiency than Cytb. Species identification using COII sequences had a higher frequency of success (96.9% in "best match" and 90.8% in "best close match") and yielded lower intra- and higher interspecific genetic divergence values than the other two markers. The use of "tag barcodes" is a new approach that involves attaching a species-specific tag to the standard DNA barcode. With this method, the "barcoding overlap" can be nearly eliminated. As a result, we were able to increase the identification success rate from 83.9% to 95.2% by using COI and the "best close match" technique.
Conclusions/significance: A COII-based identification system should be more effective in identifying lachnine species than COI or Cytb. However, the Cytb gene is an effective marker for the study of aphid population genetics due to its high sequence diversity. Furthermore, the use of "tag barcodes" can improve the accuracy of DNA barcoding identification by reducing or removing the overlap between intra- and inter-specific genetic divergence values.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures

A, B, and C are the COI, COII and Cytb NJ trees, respectively. The number of identical samples is given in brackets.

A, B, and C are the COI, COII and Cytb NJ trees, respectively.
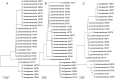
A, B, and C are the COI, COII and Cytb NJ trees, respectively.
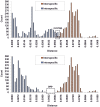
Genetic divergence was calculated using K2P model.
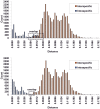
Genetic divergence was calculated using K2P model.
Similar articles
-
Barcoding aphids (Hemiptera: Aphididae) of the Korean Peninsula: updating the global data set.
Lee W, Kim H, Lim J, Choi HR, Kim Y, Kim YS, Ji JY, Foottit RG, Lee S. Lee W, et al. Mol Ecol Resour. 2011 Jan;11(1):32-7. doi: 10.1111/j.1755-0998.2010.02877.x. Mol Ecol Resour. 2011. PMID: 21429098
-
Laopichienpong N, Muangmai N, Supikamolseni A, Twilprawat P, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. Laopichienpong N, et al. Gene. 2016 Dec 15;594(2):238-247. doi: 10.1016/j.gene.2016.09.017. Epub 2016 Sep 12. Gene. 2016. PMID: 27632899
-
Kinyanjui G, Khamis FM, Mohamed S, Ombura LO, Warigia M, Ekesi S. Kinyanjui G, et al. Bull Entomol Res. 2016 Feb;106(1):63-72. doi: 10.1017/S0007485315000796. Epub 2015 Oct 22. Bull Entomol Res. 2016. PMID: 26490301
-
DNA barcode markers applied to seafood authentication: an updated review.
Fernandes TJR, Amaral JS, Mafra I. Fernandes TJR, et al. Crit Rev Food Sci Nutr. 2021;61(22):3904-3935. doi: 10.1080/10408398.2020.1811200. Epub 2020 Aug 25. Crit Rev Food Sci Nutr. 2021. PMID: 32838560 Review.
-
20 years since the introduction of DNA barcoding: from theory to application.
Fišer Pečnikar Ž, Buzan EV. Fišer Pečnikar Ž, et al. J Appl Genet. 2014 Feb;55(1):43-52. doi: 10.1007/s13353-013-0180-y. Epub 2013 Nov 8. J Appl Genet. 2014. PMID: 24203863 Review.
Cited by
-
Nibouche S, Costet L, Medina RF, Holt JR, Sadeyen J, Zoogones AS, Brown P, Blackman RL. Nibouche S, et al. PLoS One. 2021 Mar 25;16(3):e0241881. doi: 10.1371/journal.pone.0241881. eCollection 2021. PLoS One. 2021. PMID: 33764987 Free PMC article.
-
Huang YF, Bozdoğan H, Chen TH, Hsieh CC, Nai YS. Huang YF, et al. Mitochondrial DNA B Resour. 2022 Jan 18;7(1):219-221. doi: 10.1080/23802359.2021.2023333. eCollection 2022. Mitochondrial DNA B Resour. 2022. PMID: 35071762 Free PMC article.
-
Coeur d'Acier A, Cruaud A, Artige E, Genson G, Clamens AL, Pierre E, Hudaverdian S, Simon JC, Jousselin E, Rasplus JY. Coeur d'Acier A, et al. PLoS One. 2014 Jun 4;9(6):e97620. doi: 10.1371/journal.pone.0097620. eCollection 2014. PLoS One. 2014. PMID: 24896814 Free PMC article.
-
Adeoba MI, Kabongo R, der Bank HV, Yessoufou K. Adeoba MI, et al. Zookeys. 2018 Mar 26;(746):105-121. doi: 10.3897/zookeys.746.13502. eCollection 2018. Zookeys. 2018. PMID: 29674898 Free PMC article.
-
Kanturski M, Lee Y, Choi J, Lee S. Kanturski M, et al. Sci Rep. 2018 Jun 13;8(1):8998. doi: 10.1038/s41598-018-27218-2. Sci Rep. 2018. PMID: 29899412 Free PMC article.
References
-
- McKenzie M, Chiotis M, Pinkert CA, Trounce IA (2003) Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Molecular Biology and Evolution 20: 1117–1124. - PubMed
-
- Cognato AI (2006) Standard percent DNA sequence difference for insects does not predict species boundaries. Journal of Economic Entomology 99: 1037–1045. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by the National Natural Sciences Foundation of China (No. 30830017), National Science Funds for Distinguished Young Scientists (No. 31025024), National Science Fund for Fostering Talents in Basic Research (No. J0930004), and the Ministry of Science and Technology of the People's Republic of China (MOST Grant No. 2011FY120200). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Research Materials