Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus - PubMed
Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus
Nathalie Lejal et al. Antimicrob Agents Chemother. 2013 May.
Abstract
The nucleoprotein (NP) binds the viral RNA genome and associates with the polymerase in a ribonucleoprotein complex (RNP) required for transcription and replication of influenza A virus. NP has no cellular counterpart, and the NP sequence is highly conserved, which led to considering NP a hot target in the search for antivirals. We report here that monomeric nucleoprotein can be inhibited by a small molecule binding in its RNA binding groove, resulting in a novel antiviral against influenza A virus. We identified naproxen, an anti-inflammatory drug that targeted the nucleoprotein to inhibit NP-RNA association required for NP function, by virtual screening. Further docking and molecular dynamics (MD) simulations identified in the RNA groove two NP-naproxen complexes of similar levels of interaction energy. The predicted naproxen binding sites were tested using the Y148A, R152A, R355A, and R361A proteins carrying single-point mutations. Surface plasmon resonance, fluorescence, and other in vitro experiments supported the notion that naproxen binds at a site identified by MD simulations and showed that naproxen competed with RNA binding to wild-type (WT) NP and protected active monomers of the nucleoprotein against proteolytic cleavage. Naproxen protected Madin-Darby canine kidney (MDCK) cells against viral challenges with the H1N1 and H3N2 viral strains and was much more effective than other cyclooxygenase inhibitors in decreasing viral titers of MDCK cells. In a mouse model of intranasal infection, naproxen treatment decreased the viral titers in mice lungs. In conclusion, naproxen is a promising lead compound for novel antivirals against influenza A virus that targets the nucleoprotein in its RNA binding groove.
Figures
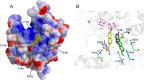
(A) Binding of naproxen to the RNA binding groove of the nucleoprotein from influenza A H1N1 virus based on PDB 2IQH (5). The protein surface is shown according to the electrostatic potential (blue, positive potential arising from the multiple arginine and lysine residues in the RNA binding groove; red, negative potential). This structure of the NP-naproxen complex solvated by water molecules was obtained after 10 ns of MD simulations. Details of the interactions of naproxen with NP inserted in a small hydrophobic cavity defined by Y148 and F489 are shown in panel B. (B) Superposition of the NP-naproxen complexes with the lowest energy. Virtual screening was used to define the possible binding site(s) of naproxen in the RNA binding groove of NP. The most MD-stable structure of the initial naproxen structure (whose carbon atoms are colored in green) is stabilized by hydrophobic interactions with Y148 and F489, a salt bridge with R361, and a water-mediated salt bridge with R355. Y148 stacks on the naphthalene core of naproxen, and the methoxy group of naproxen often forms an H-bond interaction with Q149 (see also Fig. S1 and Movie S6 in the supplemental material). The naproxen was further docked on NP using a searching volume extended to the rigid RNA binding site. The docking poses are represented as purple dots. The naproxen structure (with carbon atoms colored in yellow) with the lowest interaction energy (electrostatic and van der Waals) made a salt bridge with R152, a water-mediated salt bridge with R150, and a π-π interaction with Y148.
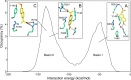
Interaction energy of the NP-naproxen complexes: the energy corresponding to interaction between naproxen and NP during the MD simulations revealed a bimodal behavior. A salt bridge between the carboxylate of naproxen and R361 shifted the naproxen from basin I (Eb1 = −55 ± 13 kcal/mol [inset A]) to basin II (Eb2 = −130 ± 14 kcal/mol [inset B]). The docking of naproxen in the extended RNA binding site generated structures with minimum interaction energy (see Fig. 1B) lying in basin II (inset C).

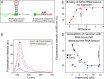
Naproxen binding to NP in competition with RNA shown by surface plasmon resonance experiments. (A) Comparison of the NP-RNA complexes formed in the absence (solid black line) and in the presence of 10 μM naproxen (dashed blue line). In this experiment, the RNA fragment was attached via its biotin 5′ extremity to surface-bound streptavidin. (B) Same experiment as described for panel A performed with the R361A mutant (solid black and dashed red lines), showing that naproxen could not compete with RNA binding to this mutant; therefore, R361 is required for the competitive binding of naproxen to NP. In contrast, naproxen competed with RNA binding to R152A (full dark greens and dotted orange lines), showing that naproxen binding is not extensively affected by mutation of this residue. (C) Binding of naproxen (1 μM) to surface-bound NP (blue trace, left scale). Note the absence of binding of naproxen (1 μM) to R361A (red trace, left scale). Both surface-bound proteins remained active and could bind RNA (dashed blue and red lines, right scale [NP and R361A, respectively]). (D) Dose dependence of naproxen binding to NP (300 nM) in competition with RNA. The maximum RU (RUmax) and RU are the signals observed with NP in the absence and presence of naproxen, respectively. Data are the means of the results of three experiments with standard deviations and could be fitted by a simple binding isotherm, yielding IC50 = 1.5 ± 0.5 μM.
Fluorescence assay of naproxen binding to NP. (A) Schematic principle of the assay using a fluorescent RNA hairpin labeled at its 5′ end with Cy3 fluorescent dye and at its 3′ end with a quencher (called an RNA beacon). (B) Fluorescence spectrum of the free beacon and in the presence of increasing NP concentrations. (C) Binding of NP to the RNA beacon monitored at 560 nm. In the free beacon, the proximity of the 3′ and 5′ ends in the paired hairpin quenched the Cy3 fluorescence due to the proximity of the quencher. Because NP preferentially binds to linear single-stranded RNA (52), NP binding to the RNA hairpin dissociates its stem (consecutive fluorescence enhancements are shown in panels B and C). These curves allow a quantitative determination of the Ki of NP for RNA. (D) Naproxen binding to NP competes with the dissociation of the hairpin stem; this is not the case in the R361A mutant. In this experiment, naproxen was added to highly fluorescent RNA-bound NP complex. Naproxen competed with RNA binding to NP and released free RNA beacon, which folded back to form the initial paired hairpin; thus, the fluorescence level decreased (black squares). Under the same conditions, the R361A mutant formed a weaker complex with the RNA beacon, as reflected by the lower initial fluorescence intensity, in agreement with previous results (20). Naproxen binding to R361A was impeded by this mutation, in agreement with the lack of fluorescence change seen upon addition of naproxen (blue circles). The values shown in panels C and D represent the results of 1 of 2 experiments with standard errors (SE).
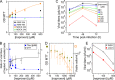
Effects of naproxen on cell viability and viral titers in the absence or presence of influenza A virus. (A) MTT tests. Data represent levels of viability (± SE) of noninfected A549 cells (black squares and blue triangles) and MDCK cells (brown triangles and green open circles) in the presence of various concentrations of naproxen added to the cell supernatants at t = 0 after incubation for 24 or 48 h (or 72 h; see Table 1). (B) Left scale (blue squares), ratio (R) of titers of treated cells (t) to titers of untreated infected cells (t0), R = t/t0; right scale (open black circles), results (± SE) of MTT viability tests of MDCK cells infected with IAV at MOI = 10−3 in the presence or absence of different naproxen concentrations at 48 h postinfection (p.i.). The experiments whose results are shown in panels A to D were performed under the same culture conditions. (C) Kinetics of viral titers of MDCK cells infected with influenza A/WSN/33 virus at MOI = 10−3. A large decrease of the titers was observed in the presence of increasing naproxen concentrations indicated as follows: 15 μM, blue circles; 50 μM, green triangles; 100 μM, gray triangles; 250 μM, brown diamonds; 500 μM, red triangles (compared to the infected and nontreated controls for [naproxen] = 0 [black squares] at 24, 48, and 72 h postinfection). The values shown in panel C are the means of the results of 4 experiments with standard deviations. (D) Toxicity assays for CC50 of naproxen determinations (see also Table 1). Left scale (gray circles), MTT tests; right scale (brown open squares), trypan blue test. Data represent results ± SE. The lines in panels B and D correspond to the fit of the data to a dose-response curve. (E) Similar levels of protection by naproxen treatment of MDCK cells challenged by two viral strains of influenza A virus, WSN/33 (H1N1) and UDORN/72 (H3N2), 24 h p.i. at MOI = 10−2.
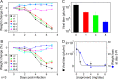
In vivo protection by naproxen treatment of mice infected with influenza A/PR8 virus loads of 2,000 PFU. (A and B) Weight change of infected mice with or without naproxen treatment administered via IP injection at the specified doses (in mg/mouse/day) to mice I-0, I-1, I-3, I-4, and I-8 (0, 1, 3, 4, and 8 mg naproxen, respectively) and the matched uninfected controls (NI-0, NI-1, NI-3, NI-4, and NI-8, respectively) at the same naproxen concentration. Each panel represents a single experiment of n = 3 experiments performed. A total of 7 to 10 mice per naproxen concentration were used in each experiment. The data are expressed as median values of the weight change in percent ± variance. Statistics analysis with the paired t test revealed meaningful differences (P being below 0.05) between I-0 and I-1 or I-3 or I-4 or I-8 and between I-1 and I-3. (B) No statistical differences between the data for I-4 and I-8 were seen. (C) Decrease of the viral titers in mice lungs as a function of the naproxen dose; statistically significant differences were found between the lung viral titer with no naproxen treatment and each of the naproxen concentrations, namely, I-0 versus I-1 (P < 0.01) and I-0 versus I-3 and I-0 versus I-8 (P < 0.005, respectively). Under these conditions, the decrease of the viral titer seen after administration of 0.2 mg oseltamivir was ca. 100-fold compared to 42-fold with 2 doses of 4 mg naproxen. (D) Comparison of the weight loss decrease and the viral titer decrease values as a function of the naproxen concentration; the fits determined by a dose-response model yielded quite similar values of EC50 = 1.6 ± 0.5 and 0.85 ± 0.42 mg/day, respectively. In this figure and Fig. 7, the data are expressed as the medians and variances of the weight changes at day i = 100 × (w0 − wi)/w0, where wi and w0 are the weights at day i and 0 postinfection calculated for each condition for infected and uninfected mice at each naproxen dose.
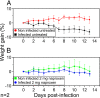
In vivo protection by naproxen treatment of mice infected with influenza A/PR8 virus loads of 50 PFU. (A) Weight change of mice infected with 50 PFU IAV and its uninfected control. (B) Same experiment as described for panel A, with mice treated by intranasal administration of naproxen at a dose of 2 mg/day. Note that both naproxen and oseltamivir (not shown) completely inhibited the weight loss in the infected untreated animals. There were statistical differences between the untreated and treated mice (I-0 and I-2 [A and B]) and the infected and noninfected untreated mice (NI-0 and I-0 [A]).
Similar articles
-
Tarus B, Bertrand H, Zedda G, Di Primo C, Quideau S, Slama-Schwok A. Tarus B, et al. J Biomol Struct Dyn. 2015 Sep;33(9):1899-912. doi: 10.1080/07391102.2014.979230. Epub 2014 Nov 19. J Biomol Struct Dyn. 2015. PMID: 25333630 Free PMC article.
-
Dilly S, Fotso Fotso A, Lejal N, Zedda G, Chebbo M, Rahman F, Companys S, Bertrand HC, Vidic J, Noiray M, Alessi MC, Tarus B, Quideau S, Riteau B, Slama-Schwok A. Dilly S, et al. J Med Chem. 2018 Aug 23;61(16):7202-7217. doi: 10.1021/acs.jmedchem.8b00557. Epub 2018 Aug 3. J Med Chem. 2018. PMID: 30028133
-
Terrier O, Dilly S, Pizzorno A, Chalupska D, Humpolickova J, Bouřa E, Berenbaum F, Quideau S, Lina B, Fève B, Adnet F, Sabbah M, Rosa-Calatrava M, Maréchal V, Henri J, Slama-Schwok A. Terrier O, et al. Molecules. 2021 Apr 29;26(9):2593. doi: 10.3390/molecules26092593. Molecules. 2021. PMID: 33946802 Free PMC article.
-
Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target.
Chenavas S, Crépin T, Delmas B, Ruigrok RW, Slama-Schwok A. Chenavas S, et al. Future Microbiol. 2013 Dec;8(12):1537-45. doi: 10.2217/fmb.13.128. Future Microbiol. 2013. PMID: 24266354 Review.
-
Influenza nucleoprotein: promising target for antiviral chemotherapy.
Cianci C, Gerritz SW, Deminie C, Krystal M. Cianci C, et al. Antivir Chem Chemother. 2012 Jul 26;23(3):77-91. doi: 10.3851/IMP2235. Antivir Chem Chemother. 2012. PMID: 22837443 Review.
Cited by
-
Stevaert A, Naesens L. Stevaert A, et al. Med Res Rev. 2016 Nov;36(6):1127-1173. doi: 10.1002/med.21401. Epub 2016 Aug 29. Med Res Rev. 2016. PMID: 27569399 Free PMC article. Review.
-
The role of adjuvant immunomodulatory agents for treatment of severe influenza.
Hui DS, Lee N, Chan PK, Beigel JH. Hui DS, et al. Antiviral Res. 2018 Feb;150:202-216. doi: 10.1016/j.antiviral.2018.01.002. Epub 2018 Jan 8. Antiviral Res. 2018. PMID: 29325970 Free PMC article. Review.
-
Tarus B, Bertrand H, Zedda G, Di Primo C, Quideau S, Slama-Schwok A. Tarus B, et al. J Biomol Struct Dyn. 2015 Sep;33(9):1899-912. doi: 10.1080/07391102.2014.979230. Epub 2014 Nov 19. J Biomol Struct Dyn. 2015. PMID: 25333630 Free PMC article.
-
Chang CK, Lo SC, Wang YS, Hou MH. Chang CK, et al. Drug Discov Today. 2016 Apr;21(4):562-72. doi: 10.1016/j.drudis.2015.11.015. Epub 2015 Dec 12. Drug Discov Today. 2016. PMID: 26691874 Free PMC article. Review.
-
Hu Y, Sneyd H, Dekant R, Wang J. Hu Y, et al. Curr Top Med Chem. 2017;17(20):2271-2285. doi: 10.2174/1568026617666170224122508. Curr Top Med Chem. 2017. PMID: 28240183 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous