Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs - PubMed
- ️Tue Jan 01 2013
. 2013 Jul 18;154(2):285-96.
doi: 10.1016/j.cell.2013.06.031.
Tom Shemesh, Narayanan Kasthuri, Robin W Klemm, Richard Schalek, Kenneth J Hayworth, Arthur R Hand, Maya Yankova, Greg Huber, Jeff W Lichtman, Tom A Rapoport, Michael M Kozlov
Affiliations
- PMID: 23870120
- PMCID: PMC3767119
- DOI: 10.1016/j.cell.2013.06.031
Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs
Mark Terasaki et al. Cell. 2013.
Abstract
The endoplasmic reticulum (ER) often forms stacked membrane sheets, an arrangement that is likely required to accommodate a maximum of membrane-bound polysomes for secretory protein synthesis. How sheets are stacked is unknown. Here, we used improved staining and automated ultrathin sectioning electron microscopy methods to analyze stacked ER sheets in neuronal cells and secretory salivary gland cells of mice. Our results show that stacked ER sheets form a continuous membrane system in which the sheets are connected by twisted membrane surfaces with helical edges of left- or right-handedness. The three-dimensional structure of tightly stacked ER sheets resembles a parking garage, in which the different levels are connected by helicoidal ramps. A theoretical model explains the experimental observations and indicates that the structure corresponds to a minimum of elastic energy of sheet edges and surfaces. The structure allows the dense packing of ER sheets in the restricted space of a cell.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
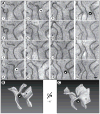
(A) Membranes were visualized in serial sections, each 30 nm thick, by scanning EM at 3 nm per pixel. The order of the sections is indicated by the numbers. Supplemental Figure S1 shows an Image J stack of the sections. A region of cytoplasm containing only ER sheets (i.e., no tubules) is shown. Two instances of the common connection motif are shown: between sheets ‘a’ and ‘b’ and between sheets ‘b’ and ‘c’; they are highlighted by arrowheads in circles of white and black background, respectively. The arrowhead on black background corresponds to the one shown in the 3D reconstruction in Figures 1B and 1C, and in higher magnification in Figure 2. Scale bar = 200 nm. (B) A 3D reconstruction of the serial section series in (A). The view is in the same orientation, with the same three sheets labeled. The arrow indicates the twist of the sheet connection (according to convention, this structure is “left handed”, because the edge turns counter-clockwise as it moves into the plane of the paper). Movie S1 shows a Quicktime movie of the reconstruction being rotated. (C) As in (B), but rotated by 20°, as indicated.
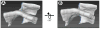
(A) Shown is a 3D reconstruction of a single sheet connection. The region corresponds to that indicated by an arrowhead on black background in Figure 1A. The orientation was preserved. Note that a sheet edge (shown as a blue transparent line) runs through the connection and is present on both sides of an apparent three-way sheet junction. Movie S2 shows a Quicktime movie of the reconstruction being rotated. (B) As in (A), but the reconstruction is rotated around the horizontal axis, as indicated.

(A) Membranes were visualized in serial sections, each 40 nm thick, by scanning EM at 3 nm per pixel. The top panel shows the region that was analyzed by serial sectioning EM. Supplemental Figure S2 shows an Image J stack of the sections. Serial sections of the regions indicated by the black and white rectangles are shown in the lower panel in the left and right columns. The sheets are labeled with letters ‘a’ to ‘d’ (black rectangle, left column) and ‘e’ to ‘j’ (white rectangle, right column). Note that the sheets break and re-attach to connect adjacent sheets of the stack (for example, in the left column, sheet ‘b’ breaks in frame 3, and rejoins with sheet ‘c’ in frame 4). Scale bar = 200 nm. (B) 3D reconstruction of the area shown in the top panel of (A). Movie S3 shows a Quicktime movie of this reconstruction being rotated.
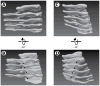
(A) A 3D reconstruction of the region within the black rectangle of Figure 3A. The sheets are labeled as in Figure 3A (left column). Movie S4 shows a Quicktime movie of this reconstruction being rotated. (B) As in (A), but from a different view angle. The arrows indicate the handedness of the helical edge. (C) A 3D reconstruction of the region within the white rectangle of Figure 3A. The sheets are labeled as in Figure 3A (right two columns). Movie S5 shows a Quicktime movie of this reconstruction being rotated. (D) As in (C), but from a different view angle. The arrows indicate the handedness of the helical edge, which is opposite to that in (A) and (B). See also Figure S3.

(A) Model for a single helicoidal connection. Shown is a section of a hollow helicoidal element with an internal radius of 75 nm and a pitch of 200 nm. The different levels of the sheet stack are shown in different colors. The helicoid surface is viewed along an axis (Z) that has an elevation angle of 50° from the axis of the helicoid. (B) Calculated serial cross sections for the helicoidal connection shown in (A). The cross-sections were computed at 30 nm intervals for sheets of 30 nm thickness. The cross sections are calculated along the Z-axis. The Z origin is arbitrary, and units are in nm. See also Figure S4.

(A) Top view of the computed optimal shape of sheets joined by helicoidal connections. The helicoidal elements of alternating handedness are arranged in a rectangular array. The surface is composed of slightly deformed helicoidal surfaces, shown in red, and saddle-like surfaces that smoothly connect them, shown in green. The surface is obtained by simultaneous minimization of the energy of the helicoidal and saddle-like elements with the constraint that the surface is continuous across their interface. The helical edges have a pitch of 220 nm and a radius of 65 nm. (B) Side views of a 3D model of the optimized structure, a sub-region of the surface in (A). Note the similarity with a parking garage. Movie S7 shows a Quicktime movie of this structure being rotated. (C) Calculated serial cross-sections obtained for the optimal surface shown in (A) and (B), with a 30 nm distance between the section planes. The cross sections were calculated along an axis Z, which corresponds to the viewing direction in the right panel of (B). The origin is arbitrary and the units are in nm. See also Figure S4 and Figure S5.
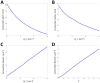
(A) Optimized structures of stacked sheets with helicoidal connections were calculated. The pitch of the helicoidal connections was determined for different values of the absolute value of the edge spontaneous curvature, |ζs|, (ζs < 0).The calculations were performed for a lateral pressure Π = 4 κ μm−2 (where κ is the sheet plane bending rigidity), and a normalized dimensionless edge length Γ = 6. (B) As in (A), but the edge radius was plotted versus |ζs|. (C) As in (A), but the connection density was plotted versus |ζs |. (D) The connections density was calculated as a function of the normalized edge length, Γ. The calculations were performed for ζs = −10 μm−1 and Π = 4 κ μm−2.
Comment in
-
Differential geometry meets the cell.
Marshall WF. Marshall WF. Cell. 2013 Jul 18;154(2):265-6. doi: 10.1016/j.cell.2013.06.032. Cell. 2013. PMID: 23870115
Similar articles
-
Differential geometry meets the cell.
Marshall WF. Marshall WF. Cell. 2013 Jul 18;154(2):265-6. doi: 10.1016/j.cell.2013.06.032. Cell. 2013. PMID: 23870115
-
Mechanisms determining the morphology of the peripheral ER.
Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Shibata Y, et al. Cell. 2010 Nov 24;143(5):774-88. doi: 10.1016/j.cell.2010.11.007. Cell. 2010. PMID: 21111237 Free PMC article.
-
Form follows function: the importance of endoplasmic reticulum shape.
Westrate LM, Lee JE, Prinz WA, Voeltz GK. Westrate LM, et al. Annu Rev Biochem. 2015;84:791-811. doi: 10.1146/annurev-biochem-072711-163501. Epub 2015 Jan 12. Annu Rev Biochem. 2015. PMID: 25580528 Review.
-
Endoplasmatic reticulum shaping by generic mechanisms and protein-induced spontaneous curvature.
Sackmann E. Sackmann E. Adv Colloid Interface Sci. 2014 Jun;208:153-60. doi: 10.1016/j.cis.2014.02.006. Epub 2014 Feb 15. Adv Colloid Interface Sci. 2014. PMID: 24607001 Review.
Cited by
-
Wang ZH, Rabouille C, Geisbrecht ER. Wang ZH, et al. Biol Open. 2015 Apr 10;4(5):636-48. doi: 10.1242/bio.201511551. Biol Open. 2015. PMID: 25862246 Free PMC article.
-
Nguyen HB, Thai TQ, Sui Y, Azuma M, Fujiwara K, Ohno N. Nguyen HB, et al. Front Neural Circuits. 2018 Nov 23;12:108. doi: 10.3389/fncir.2018.00108. eCollection 2018. Front Neural Circuits. 2018. PMID: 30532696 Free PMC article. Review.
-
Roles for the Endoplasmic Reticulum in Regulation of Neuronal Calcium Homeostasis.
Karagas NE, Venkatachalam K. Karagas NE, et al. Cells. 2019 Oct 10;8(10):1232. doi: 10.3390/cells8101232. Cells. 2019. PMID: 31658749 Free PMC article. Review.
-
Lazutin AA, Vasilevskaya VV. Lazutin AA, et al. ACS Omega. 2018 Oct 10;3(10):12967-12974. doi: 10.1021/acsomega.8b01643. eCollection 2018 Oct 31. ACS Omega. 2018. PMID: 31458020 Free PMC article.
-
Axonal Endoplasmic Reticulum Dynamics and Its Roles in Neurodegeneration.
Öztürk Z, O'Kane CJ, Pérez-Moreno JJ. Öztürk Z, et al. Front Neurosci. 2020 Jan 29;14:48. doi: 10.3389/fnins.2020.00048. eCollection 2020. Front Neurosci. 2020. PMID: 32116502 Free PMC article. Review.
References
-
- Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources