miRBase: annotating high confidence microRNAs using deep sequencing data - PubMed
. 2014 Jan;42(Database issue):D68-73.
doi: 10.1093/nar/gkt1181. Epub 2013 Nov 25.
Affiliations
- PMID: 24275495
- PMCID: PMC3965103
- DOI: 10.1093/nar/gkt1181
miRBase: annotating high confidence microRNAs using deep sequencing data
Ana Kozomara et al. Nucleic Acids Res. 2014 Jan.
Abstract
We describe an update of the miRBase database (http://www.mirbase.org/), the primary microRNA sequence repository. The latest miRBase release (v20, June 2013) contains 24 521 microRNA loci from 206 species, processed to produce 30 424 mature microRNA products. The rate of deposition of novel microRNAs and the number of researchers involved in their discovery continue to increase, driven largely by small RNA deep sequencing experiments. In the face of these increases, and a range of microRNA annotation methods and criteria, maintaining the quality of the microRNA sequence data set is a significant challenge. Here, we describe recent developments of the miRBase database to address this issue. In particular, we describe the collation and use of deep sequencing data sets to assign levels of confidence to miRBase entries. We now provide a high confidence subset of miRBase entries, based on the pattern of mapped reads. The high confidence microRNA data set is available alongside the complete microRNA collection at http://www.mirbase.org/. We also describe embedding microRNA-specific Wikipedia pages on the miRBase website to encourage the microRNA community to contribute and share textual and functional information.
Figures

The patterns of small RNA deep sequencing reads mapping to three mouse microRNAs. Hairpin microRNA sequences are shown at the bottom of each panel, with derived mature microRNA sequences shown in magenta, and predicted base-paired secondary structure in dot-bracket notation underneath. Read sequences are shown in blue, with the summed count across all data sets on the right. (A) The annotation of mmu-mir-3072 is supported by reads mapped to both mature sequences, pairing with a 2-nt 3′ overhang, and mmu-mir-3072 is therefore annotated as a high confidence microRNA. (B) Reads from the available deep sequencing data sets map to only one arm of the mmu-mir-184 hairpin, which cannot therefore be annotated with high confidence. (C) The pattern of reads mapping to the annotated mmu-mir-1940 locus refutes the microRNA annotation—reads mapping to the two arms of the predicted hairpin do not pair with the 2-nt 3′ overhang characteristic of microRNA processing.
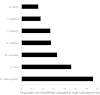
Proportions of microRNAs in model species that are included in the high confidence microRNA set.
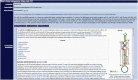
The miRBase entry page for dme-mir-10, showing the embedded Wikipedia page on the mir-10 microRNA precursor family.
Similar articles
-
miRBase: integrating microRNA annotation and deep-sequencing data.
Kozomara A, Griffiths-Jones S. Kozomara A, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D152-7. doi: 10.1093/nar/gkq1027. Epub 2010 Oct 30. Nucleic Acids Res. 2011. PMID: 21037258 Free PMC article.
-
miRBase: from microRNA sequences to function.
Kozomara A, Birgaoanu M, Griffiths-Jones S. Kozomara A, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D155-D162. doi: 10.1093/nar/gky1141. Nucleic Acids Res. 2019. PMID: 30423142 Free PMC article.
-
miRBase: the microRNA sequence database.
Griffiths-Jones S. Griffiths-Jones S. Methods Mol Biol. 2006;342:129-38. doi: 10.1385/1-59745-123-1:129. Methods Mol Biol. 2006. PMID: 16957372 Review.
-
miRBase: microRNA sequences and annotation.
Griffiths-Jones S. Griffiths-Jones S. Curr Protoc Bioinformatics. 2010 Mar;Chapter 12:12.9.1-12.9.10. doi: 10.1002/0471250953.bi1209s29. Curr Protoc Bioinformatics. 2010. PMID: 20205188
-
Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. Fromm B, et al. Annu Rev Genet. 2015;49:213-42. doi: 10.1146/annurev-genet-120213-092023. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473382 Free PMC article. Review.
Cited by
-
Roles of microRNAs in Regulating Apoptosis in the Pathogenesis of Endometriosis.
Azam INA, Wahab NA, Mokhtar MH, Shafiee MN, Mokhtar NM. Azam INA, et al. Life (Basel). 2022 Aug 26;12(9):1321. doi: 10.3390/life12091321. Life (Basel). 2022. PMID: 36143357 Free PMC article. Review.
-
Yin H, Zhao J, He H, Chen Y, Wang Y, Li D, Zhu Q. Yin H, et al. Int J Mol Sci. 2020 Aug 4;21(15):5573. doi: 10.3390/ijms21155573. Int J Mol Sci. 2020. PMID: 32759823 Free PMC article.
-
Li J, Lei K, Wu Z, Li W, Liu G, Liu J, Cheng F, Tang Y. Li J, et al. Oncotarget. 2016 Jul 19;7(29):45584-45596. doi: 10.18632/oncotarget.10052. Oncotarget. 2016. PMID: 27329603 Free PMC article.
-
BmncRNAdb: a comprehensive database of non-coding RNAs in the silkworm, Bombyx mori.
Zhou QZ, Zhang B, Yu QY, Zhang Z. Zhou QZ, et al. BMC Bioinformatics. 2016 Sep 13;17(1):370. doi: 10.1186/s12859-016-1251-y. BMC Bioinformatics. 2016. PMID: 27623959 Free PMC article.
-
Sheikh AM, Small HY, Currie G, Delles C. Sheikh AM, et al. PLoS One. 2016 Aug 16;11(8):e0160808. doi: 10.1371/journal.pone.0160808. eCollection 2016. PLoS One. 2016. PMID: 27529341 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources