A customized Web portal for the genome of the ctenophore Mnemiopsis leidyi - PubMed
- ️Wed Jan 01 2014
A customized Web portal for the genome of the ctenophore Mnemiopsis leidyi
R Travis Moreland et al. BMC Genomics. 2014.
Abstract
Background: Mnemiopsis leidyi is a ctenophore native to the coastal waters of the western Atlantic Ocean. A number of studies on Mnemiopsis have led to a better understanding of many key biological processes, and these studies have contributed to the emergence of Mnemiopsis as an important model for evolutionary and developmental studies. Recently, we sequenced, assembled, annotated, and performed a preliminary analysis on the 150-megabase genome of the ctenophore, Mnemiopsis. This sequencing effort has produced the first set of whole-genome sequencing data on any ctenophore species and is amongst the first wave of projects to sequence an animal genome de novo solely using next-generation sequencing technologies.
Description: The Mnemiopsis Genome Project Portal (http://research.nhgri.nih.gov/mnemiopsis/) is intended both as a resource for obtaining genomic information on Mnemiopsis through an intuitive and easy-to-use interface and as a model for developing customized Web portals that enable access to genomic data. The scope of data available through this Portal goes well beyond the sequence data available through GenBank, providing key biological information not available elsewhere, such as pathway and protein domain analyses; it also features a customized genome browser for data visualization.
Conclusions: We expect that the availability of these data will allow investigators to advance their own research projects aimed at understanding phylogenetic diversity and the evolution of proteins that play a fundamental role in metazoan development. The overall approach taken in the development of this Web site can serve as a viable model for disseminating data from whole-genome sequencing projects, framed in a way that best-serves the specific needs of the scientific community.
Figures
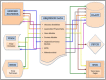
A visual representation of the Mnemiopsis Genome Project (MGP) Portal depicting the flow of Mnemiopsis sequence data into accessible internal data visualization and annotation tools. Colored arrows correspond to individual data types listed in the Sequence Data box (center). For example, the flow of ‘Assembled Transcripts’ data (e.g., Cufflinks- and Trinity-assembled RNA-seq transcripts) is represented by the orange arrows; these data can be viewed both as tracks in the Genome Browser and/or utilized as a BLAST database.
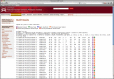
The Mnemiopsis BLAST implementation provides users an intuitive Web interface for performing sequence similarity searches. Shown are tab-delimited BLASTP results from a single human PAX3 protein, providing links to relevant sequence entries in the Mnemiopsis Genome Browser (purple ‘B’ box), individual Gene Wiki pages (green ‘G’ box), and Unfiltered Protein Models (orange ‘U’ box).
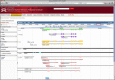
Several customized tracks can be displayed on the Mnemiopsis genome browser, implemented in JBrowse. Here, we present a predicted protein model (ML000129a) containing G-protein-coupled receptor domains, as evidenced by the PFAM2.2 track. Transcripts assembled from RNA-seq reads support the predicted protein model as depicted in the CL2 track. Transcripts are presented as gray arrows indicating the orientation of the transcript. Exons are presented as light-colored solid bars and untranslated regions (both 5’ and 3’) are darker-shaded bars. Assembled genomic scaffolds (SCF) are depicted as solid black tracks with intermittent gaps shaded bright pink. The MASK track also appears as a solid black bar highlighted with blue in the genomic regions that have been repeat-masked using VMatch. Other tracks shown include an EST and several unfiltered protein prediction models (2.2UF). Additional tracks are described in the main text.
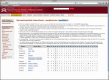
Human KEGG pathways containing genes with a Mnemiopsis homolog are presented as pathway-specific ortholog cluster matrices. Each row in the glycosphingolipid biosynthesis pathway represents a cluster from our clustering analysis. The ‘Cluster’ column indicates the most inclusive clade that encompasses all of the proteins in the cluster. The ‘Ratio’ column represents the number of human proteins in a given cluster that are found in the pathway over the total number of human proteins in that cluster. Mnemiopsis (Ml) entries are shaded in gray and hyperlinked to their respective Gene Wiki pages.
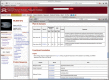
Each record in the Gene Wiki represents a single Mnemiopsis gene (ML000127a) and provides the following annotation: nucleotide and protein sequences, coding exonic genomic coordinates, and pre-computed BLAST hits from numerous organisms displaying the top hits for each protein. The inset illustrates additional annotations available through the Gene Wiki pages, including those regarding Pfam-A domains and GO terminology (functional annotation).
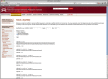
The Mnemiopsis Fetch Tool is used to retrieve a single or partial scaffold, its reverse complement or the six-frame protein translation. Displayed above is the output of a queried partial genomic scaffold for ML0001 showing the specified genomic region of interest and its six-frame protein translations (the first five translations are depicted).
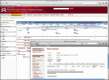
Selecting the Genome Browser link (purple ‘B’ box from Figure 1) from a BLASTP result entry of queried known homeodomains against the complete Mnemiopsis Unfiltered Protein Models directs the user to the Genome Browser displaying the applicable Mnemiopsis transcript model. Shown above the 2.2UF track in the browser is the PFAM2.2 track displaying evidence of a homeobox in the targeted region. Clicking on the ‘Homeobox’ link in the PFAM2.2 track opens a new browser window displaying the Pfam-A domain (ML1991_pfa) prediction results derived from a pre-compiled hmmscan run using HMMER. This Pfam-A domain record provides the genomic location of the Pfam-A domain, the genomic coordinates, transcript and protein sequences, and the hmmscan output for the homeodomain prediction.
Similar articles
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.
Edgar A, Mitchell DG, Martindale MQ. Edgar A, et al. Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
-
Moreland RT, Nguyen AD, Ryan JF, Baxevanis AD. Moreland RT, et al. Database (Oxford). 2020 Jan 1;2020:baaa029. doi: 10.1093/database/baaa029. Database (Oxford). 2020. PMID: 32386298 Free PMC article.
-
Schnitzler CE, Pang K, Powers ML, Reitzel AM, Ryan JF, Simmons D, Tada T, Park M, Gupta J, Brooks SY, Blakesley RW, Yokoyama S, Haddock SH, Martindale MQ, Baxevanis AD. Schnitzler CE, et al. BMC Biol. 2012 Dec 21;10:107. doi: 10.1186/1741-7007-10-107. BMC Biol. 2012. PMID: 23259493 Free PMC article.
-
Moreland RT, Zhang S, Barreira SN, Ryan JF, Baxevanis AD. Moreland RT, et al. Proteomics. 2024 Aug;24(15):e2300397. doi: 10.1002/pmic.202300397. Epub 2024 Feb 8. Proteomics. 2024. PMID: 38329168 Free PMC article.
-
Presnell JS, Bubel M, Knowles T, Patry W, Browne WE. Presnell JS, et al. Nat Protoc. 2022 Aug;17(8):1868-1900. doi: 10.1038/s41596-022-00702-w. Epub 2022 Jun 13. Nat Protoc. 2022. PMID: 35697825 Review.
Cited by
-
Sasson DA, Ryan JF. Sasson DA, et al. PeerJ. 2016 Mar 24;4:e1846. doi: 10.7717/peerj.1846. eCollection 2016. PeerJ. 2016. PMID: 27042395 Free PMC article.
-
Evolution of miRNA Tailing by 3' Terminal Uridylyl Transferases in Metazoa.
Modepalli V, Moran Y. Modepalli V, et al. Genome Biol Evol. 2017 Jun 1;9(6):1547-1560. doi: 10.1093/gbe/evx106. Genome Biol Evol. 2017. PMID: 28633361 Free PMC article.
-
Davidson PL, Koch BJ, Schnitzler CE, Henry JQ, Martindale MQ, Baxevanis AD, Browne WE. Davidson PL, et al. Mol Reprod Dev. 2017 Nov;84(11):1218-1229. doi: 10.1002/mrd.22926. Epub 2017 Nov 10. Mol Reprod Dev. 2017. PMID: 29068507 Free PMC article.
-
Ladewig L, Gloy L, Langfeldt D, Pinnow N, Weiland-Bräuer N, Schmitz RA. Ladewig L, et al. Microorganisms. 2023 Aug 29;11(9):2184. doi: 10.3390/microorganisms11092184. Microorganisms. 2023. PMID: 37764028 Free PMC article.
-
Whole-Body Regeneration in the Lobate Ctenophore Mnemiopsis leidyi.
Edgar A, Mitchell DG, Martindale MQ. Edgar A, et al. Genes (Basel). 2021 Jun 5;12(6):867. doi: 10.3390/genes12060867. Genes (Basel). 2021. PMID: 34198839 Free PMC article. Review.
References
-
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, Putnam NH, Haddock SHD, Dunn CW, Wolfsberg TG, Mullikin JC, Martindale MQ, Baxevanis AD. NISC Comparative Sequencing Program. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources