TFClass: a classification of human transcription factors and their rodent orthologs - PubMed
. 2015 Jan;43(Database issue):D97-102.
doi: 10.1093/nar/gku1064. Epub 2014 Oct 31.
Affiliations
- PMID: 25361979
- PMCID: PMC4383905
- DOI: 10.1093/nar/gku1064
TFClass: a classification of human transcription factors and their rodent orthologs
Edgar Wingender et al. Nucleic Acids Res. 2015 Jan.
Abstract
TFClass aims at classifying eukaryotic transcription factors (TFs) according to their DNA-binding domains (DBDs). For this, a classification schema comprising four generic levels (superclass, class, family and subfamily) was defined that could accommodate all known DNA-binding human TFs. They were assigned to their (sub-)families as instances at two different levels, the corresponding TF genes and individual gene products (protein isoforms). In the present version, all mouse and rat orthologs have been linked to the human TFs, and the mouse orthologs have been arranged in an independent ontology. Many TFs were assigned with typical DNA-binding patterns and positional weight matrices derived from high-throughput in-vitro binding studies. Predicted TF binding sites from human gene upstream sequences are now also attached to each human TF whenever a PWM was available for this factor or one of his paralogs. TFClass is freely available at http://tfclass.bioinf.med.uni-goettingen.de/ through a web interface and for download in OBO format.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
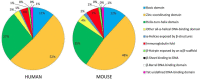
Superclass distribution of human TF genes and their mouse orthologs.

Web interface of human and mouse TFClass. In the center, the classification of human TFs is shown by default, on the right is the classification of mouse TFs. Navigating to a certain entity in the human classification (here: c-Myc) automatically opens the mouse classification to the same point. Note that the subfamily displayed here contains one human-specific factor (L-Myc-2, 1.2.6.5.4) in the central part and one mouse-specific TF (B-Myc, 1.2.6.5.6) on the right-hand side. On the left, additional information for the selected human TF is given, including external database links. The button ‘Switch classifications’ on top allows the user to put the mouse classification as primary one in the center, which would also switch the additional information on the left from human to mouse.
Similar articles
-
TFClass: expanding the classification of human transcription factors to their mammalian orthologs.
Wingender E, Schoeps T, Haubrock M, Krull M, Dönitz J. Wingender E, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D343-D347. doi: 10.1093/nar/gkx987. Nucleic Acids Res. 2018. PMID: 29087517 Free PMC article.
-
TFClass: an expandable hierarchical classification of human transcription factors.
Wingender E, Schoeps T, Dönitz J. Wingender E, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D165-70. doi: 10.1093/nar/gks1123. Epub 2012 Nov 24. Nucleic Acids Res. 2013. PMID: 23180794 Free PMC article.
-
Plant-TFClass: a structural classification for plant transcription factors.
Blanc-Mathieu R, Dumas R, Turchi L, Lucas J, Parcy F. Blanc-Mathieu R, et al. Trends Plant Sci. 2024 Jan;29(1):40-51. doi: 10.1016/j.tplants.2023.06.023. Epub 2023 Jul 22. Trends Plant Sci. 2024. PMID: 37482504 Review.
-
AnimalTFDB: a comprehensive animal transcription factor database.
Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang H, Guo AY. Zhang HM, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D144-9. doi: 10.1093/nar/gkr965. Epub 2011 Nov 12. Nucleic Acids Res. 2012. PMID: 22080564 Free PMC article.
-
Genomic repertoires of DNA-binding transcription factors across the tree of life.
Charoensawan V, Wilson D, Teichmann SA. Charoensawan V, et al. Nucleic Acids Res. 2010 Nov;38(21):7364-77. doi: 10.1093/nar/gkq617. Epub 2010 Jul 30. Nucleic Acids Res. 2010. PMID: 20675356 Free PMC article. Review.
Cited by
-
Constructing temporal regulatory cascades in the context of development and cell differentiation.
Daou R, Beißbarth T, Wingender E, Gültas M, Haubrock M. Daou R, et al. PLoS One. 2020 Apr 10;15(4):e0231326. doi: 10.1371/journal.pone.0231326. eCollection 2020. PLoS One. 2020. PMID: 32275727 Free PMC article.
-
GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments.
Yevshin I, Sharipov R, Valeev T, Kel A, Kolpakov F. Yevshin I, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D61-D67. doi: 10.1093/nar/gkw951. Epub 2016 Oct 24. Nucleic Acids Res. 2017. PMID: 27924024 Free PMC article.
-
Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, Kolpakov FA, Makeev VJ. Kulakovskiy IV, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D252-D259. doi: 10.1093/nar/gkx1106. Nucleic Acids Res. 2018. PMID: 29140464 Free PMC article.
-
Rieu P, Beretta VM, Caselli F, Thévénon E, Lucas J, Rizk M, Franchini E, Caporali E, Paleni C, Nanao MH, Kater MM, Dumas R, Zubieta C, Parcy F, Gregis V. Rieu P, et al. Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2310464121. doi: 10.1073/pnas.2310464121. Epub 2024 Feb 27. Proc Natl Acad Sci U S A. 2024. PMID: 38412122 Free PMC article.
-
SEanalysis: a web tool for super-enhancer associated regulatory analysis.
Qian FC, Li XC, Guo JC, Zhao JM, Li YY, Tang ZD, Zhou LW, Zhang J, Bai XF, Jiang Y, Pan Q, Wang QY, Li EM, Li CQ, Xu LY, Lin DC. Qian FC, et al. Nucleic Acids Res. 2019 Jul 2;47(W1):W248-W255. doi: 10.1093/nar/gkz302. Nucleic Acids Res. 2019. PMID: 31028388 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous