Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp - PubMed
- ️Thu Jan 01 2015
Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp
Jee Eun Han et al. Dis Aquat Organ. 2015.
Abstract
The 69 kb plasmid pVPA3-1 was identified in Vibrio parahaemolyticus strain 13‑028/A3 that can cause acute hepatopancreatic necrosis disease (AHPND). This disease is responsible for mass mortalities in farmed penaeid shrimp and is referred to as early mortality syndrome (EMS). The plasmid has a GC content of 45.9% with a copy number of 37 per bacterial cell as determined by comparative quantitative PCR analyses. It consists of 92 open reading frames that encode mobilization proteins, replication enzymes, transposases, virulence-associated proteins, and proteins similar to Photorhabdus insect-related (Pir) toxins. In V. parahaemolyticus, these Pir toxin-like proteins are encoded by 2 genes (pirA- and pirB-like) located within a 3.5 kb fragment flanked with inverted repeats of a transposase-coding sequence (1 kb). The GC content of these 2 genes is only 38.2%, substantially lower than that of the rest of the plasmid, which suggests that these genes were recently acquired. Based on a proteomic analysis, the pirA-like (336 bp) and pirB-like (1317 bp) genes encode for 13 and 50 kDa proteins, respectively. In laboratory cultures of V. parahaemolyticus 13-028/A3, both proteins were secreted into the culture medium. We developed a duplex PCR diagnostic method, with a detection limit of 10(5) CFU ml(-1) and targeting pirA- and pirB-like genes in this strain of V. parahaemolyticus. This PCR protocol can reliably detect AHPND-causing strains of V. parahaemolyticus and does not cross react with non-pathogenic strains or with other species of Vibrio isolated from shrimp ponds.
Figures

Plasmid pVPA3-1. All predicted open reading frames (ORFs) are shown as arrows in different colors. Direction of arrowhead represents the transcriptional orientation
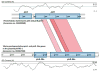
Schematic representation of the comparative analysis of the pirA- and pirB-like genes in Vibrio parahaemolyticus with the pirA and pirB genes of Photorhabdus luminescens. Regions for the pirA and pirB genes are shown with a symmetrical gene order. Translated BLAST (tblastx, score cut-off: 30) was used to align translated genome sequences. The color intensity is proportional to the sequence homology. Nucleotide base pairs are indicated between grey lines and GC contents (%) are shown

PCR detection of pirA- and pirB-like genes. (A) Amplifications with pathogenic acute hepatopancreatic necrosis disease (AHPND)-Vibrio parahaemolyticus strains and non-pathogenic strains. Pathogenic strains collected from Mexico (Lane M: 1 kb plus DNA ladder; Lane 1: strain 13-511/A1; Lane 3: strain 13-306/D4, see Table 2) and Vietnam (Lane 2: strain 13-028/A3; Lane 4: strain 12-194G); non-pathogenic strains from shrimp farms in Vietnam (Lane 5: strain 13-028/A2), India (Lane 6: strain 13-488L), and USA (Lane 7: strain 13-431). (B) Determination of sensitivity by serially diluting V. parahaemolyticus 13-028/A3 DNA; bacterial concentrations were as follows: 1 × 107 (Lane 1), 1 × 106 (Lane 2), 1 × 105 (Lane 3), 1 × 104 (Lane 4), and 1 × 103 CFU ml−1 (Lane 5), and a non-template control (Lane 6). Lane M: 1 kb plus DNA ladder
Similar articles
-
Phiwsaiya K, Charoensapsri W, Taengphu S, Dong HT, Sangsuriya P, Nguyen GTT, Pham HQ, Amparyup P, Sritunyalucksana K, Taengchaiyaphum S, Chaivisuthangkura P, Longyant S, Sithigorngul P, Senapin S. Phiwsaiya K, et al. Appl Environ Microbiol. 2017 Aug 1;83(16):e00680-17. doi: 10.1128/AEM.00680-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28576761 Free PMC article.
-
Lee CT, Chen IT, Yang YT, Ko TP, Huang YT, Huang JY, Huang MF, Lin SJ, Chen CY, Lin SS, Lightner DV, Wang HC, Wang AH, Wang HC, Hor LI, Lo CF. Lee CT, et al. Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10798-803. doi: 10.1073/pnas.1503129112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261348 Free PMC article.
-
Sirikharin R, Taengchaiyaphum S, Sanguanrut P, Chi TD, Mavichak R, Proespraiwong P, Nuangsaeng B, Thitamadee S, Flegel TW, Sritunyalucksana K. Sirikharin R, et al. PLoS One. 2015 May 27;10(5):e0126987. doi: 10.1371/journal.pone.0126987. eCollection 2015. PLoS One. 2015. PMID: 26017673 Free PMC article.
-
Lin SJ, Hsu KC, Wang HC. Lin SJ, et al. Mar Drugs. 2017 Dec 1;15(12):373. doi: 10.3390/md15120373. Mar Drugs. 2017. PMID: 29194352 Free PMC article. Review.
-
Wang HC, Lin SJ, Wang HC, Kumar R, Le PT, Leu JH. Wang HC, et al. PLoS Pathog. 2023 May 4;19(5):e1011330. doi: 10.1371/journal.ppat.1011330. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37141203 Free PMC article. Review.
Cited by
-
Shahrear S, Afroj Zinnia M, Sany MRU, Islam ABMMK. Shahrear S, et al. Bioinform Biol Insights. 2022 Nov 9;16:11779322221136002. doi: 10.1177/11779322221136002. eCollection 2022. Bioinform Biol Insights. 2022. PMID: 36386863 Free PMC article.
-
Kim YB, Park SY, Jeon HJ, Kim B, Kwon MG, Kim SM, Han JE, Kim JH. Kim YB, et al. Animals (Basel). 2024 Sep 26;14(19):2788. doi: 10.3390/ani14192788. Animals (Basel). 2024. PMID: 39409739 Free PMC article.
-
Devadas S, Bhassu S, Christie Soo TC, Yusoff FM, Shariff M. Devadas S, et al. Microbiol Resour Announc. 2018 Sep 20;7(11):e01053-18. doi: 10.1128/MRA.01053-18. eCollection 2018 Sep. Microbiol Resour Announc. 2018. PMID: 30533648 Free PMC article.
-
Experimental reproduction of White Feces Syndrome in whiteleg shrimp, Penaeus vannamei.
Aranguren Caro LF, Mai HN, Cruz-Florez R, Marcos FLA, Alenton RRR, Dhar AK. Aranguren Caro LF, et al. PLoS One. 2021 Dec 23;16(12):e0261289. doi: 10.1371/journal.pone.0261289. eCollection 2021. PLoS One. 2021. PMID: 34941926 Free PMC article.
-
Reyes G, Betancourt I, Andrade B, Panchana F, Román R, Sorroza L, Trujillo LE, Bayot B. Reyes G, et al. Front Microbiol. 2022 May 9;13:838640. doi: 10.3389/fmicb.2022.838640. eCollection 2022. Front Microbiol. 2022. PMID: 35615516 Free PMC article.
References
-
- Blackburn MB, Farrar RR, Novak NG, Lawrence SD. Remarkable susceptibility of the diamondback moth (Plutella xylostella) to ingestion of Pir toxins from Photorhabdus luminescens. Entomol Exp Appl. 2006;121:31–37.
-
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous