PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection - PubMed
- ️Fri Jan 01 2016
. 2016 Mar 24;531(7595):528-32.
doi: 10.1038/nature17184. Epub 2016 Mar 16.
Affiliations
- PMID: 26982719
- PMCID: PMC4909121
- DOI: 10.1038/nature17184
PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection
Mei T Tran et al. Nature. 2016.
Abstract
The energetic burden of continuously concentrating solutes against gradients along the tubule may render the kidney especially vulnerable to ischaemia. Acute kidney injury (AKI) affects 3% of all hospitalized patients. Here we show that the mitochondrial biogenesis regulator, PGC1α, is a pivotal determinant of renal recovery from injury by regulating nicotinamide adenine dinucleotide (NAD) biosynthesis. Following renal ischaemia, Pgc1α(-/-) (also known as Ppargc1a(-/-)) mice develop local deficiency of the NAD precursor niacinamide (NAM, also known as nicotinamide), marked fat accumulation, and failure to re-establish normal function. Notably, exogenous NAM improves local NAD levels, fat accumulation, and renal function in post-ischaemic Pgc1α(-/-) mice. Inducible tubular transgenic mice (iNephPGC1α) recapitulate the effects of NAM supplementation, including more local NAD and less fat accumulation with better renal function after ischaemia. PGC1α coordinately upregulates the enzymes that synthesize NAD de novo from amino acids whereas PGC1α deficiency or AKI attenuates the de novo pathway. NAM enhances NAD via the enzyme NAMPT and augments production of the fat breakdown product β-hydroxybutyrate, leading to increased production of prostaglandin PGE2 (ref. 5), a secreted autacoid that maintains renal function. NAM treatment reverses established ischaemic AKI and also prevented AKI in an unrelated toxic model. Inhibition of β-hydroxybutyrate signalling or prostaglandin production similarly abolishes PGC1α-dependent renoprotection. Given the importance of mitochondrial health in ageing and the function of metabolically active organs, the results implicate NAM and NAD as key effectors for achieving PGC1α-dependent stress resistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
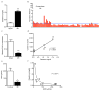
a, Serum creatinine 24 h after sham or IRI (n=5 vs. 14 mice), ***p< 0.001. b, Absence of classwide changes in intrarenal phospholipids 24 h after IRI vs. sham operation (n=6/group, NS=non-significant). Each bar represents one lipid species. P-value by two-way ANOVA. c, Renal PGC1α expression 24 h after sham or IRI (n = 5 animals per group), **p<0.01. d, Correlation of LC-MS method for serum creatinine and serum cystatin C (measured by ELISA). e, Glomerular filtration rate in controls or 24 h after IRI was determined by two-phase exponential decay curves of fluorescently-labeled inulin as described in methods (n=5/group), *p<0.05. f, Correlation of LC-MS method for serum creatinine with clearance of FITC-inulin. Curve fit according to formula sCr=κ/GFR where κ is a constant. Error bars SEM.
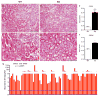
a–d, Low (a,b) and high-power (c,d) photomicrographs 24 h after IRI in WT vs. PGC1α−/− (KO) mice. Scale bars 100 and 50 μm. e,f, Blinded scoring of tubular injury in cortex and outer stripe of outer medulla (OSOM) on 4-point injury scale as described in Methods (n=8 WT vs.12 KO mice), *p<0.05. g, Di-/tri-acylglycerols (DAGs, TAGs) in renal homogenates of KO mice at baseline and 24 h after injury (n=6/group). Each bar represents one lipid species. P-value by two-way ANOVA. Error bars SEM.
a, Heatmaps (red=higher, green=lower) of Bonferroni-corrected significantly different metabolites in sham vs. IRI kidneys and WT vs. KO kidneys. Metabolites listed in purple are shared between settings. b, Total ion chromatogram of polar, positive ion mode method for representative WT-IRI sample, with niacinamide (Nam) peak at retention time = 3.88 minutes. Inset shows representative niacinamide peaks for kidney extracts from WT control (Ctrl) and WT-IRI (IRI) mice. c–e, Relative renal Nam abundance in kidneys of KO mice vs. WT littermates; WT littermates at baseline and 24 h after IRI; and KO mice at baseline and 24 h after IRI (n=6/group). f, Relative renal Nam concentrations in kidneys of mice following vehicle (Veh) vs. Nam treatment (400 mg/kg IP × 4d) with and without IRI 24 h prior to tissue collection (n=6/group). P-values by two-way ANOVA. g,h, Oil Red O stain (pink) for fat accumulation 24 h after IRI with or without Nam pretreatment (400 mg/kg IP × 4 d), scale bar 20 μm. Error bars SEM, *p<0.05, **p<0.01, ***p<0.001.
a, Schematic for generating iNephPGC1α mice. b, Relative renal PGC1α expression in controls vs. iNephPGC1α mice with and without 4 weeks of doxycycline in drinking water (n = 5/group, **p<0.01 vs. all other groups). c, Ratio of kidney weight to total body weight (note body weights statistically indistinguishable as well, n=4/group). d, Example gross images with 1 cm scale of control vs. iNephPGC1α kidney. e, Renal mitochondrial DNA (mtDNA) copy number as described in Methods. f, Relative renal gene expression of PGC1α targets (Ndufs1, Cycs, Atp5o), partnering transcription factors (Nrf1), and the mitochondrial transcription factor, TFAM. Results analyzed by two-way ANOVA with p-value for genotype as noted. N=8/group. *p<0.05 vs. control after Bonferroni correction. g, Western analysis of kidney lysates for Transcription Factor A, Mitochondrial (TFAM) and loading control. h,i, Transmission EM of mitochondria sectioned perpendicular and parallel to long axis demonstrating normal morphology in iNephPGC1α mice (representative of n=4/group), scale bar 500 nm. j,k, Blinded scoring of tubular injury in cortex and outer stripe of outer medulla (n=8 control; 12 iNephPGC1α). Error bars SEM, *p<0.05, **p<0.01.
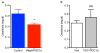
a, Serum creatinine 24 h after bacterial endotoxin injection (LPS O111:B4), n=9/group. b, Serum creatinine 24 h after bacterial endotoxin (LPS O111:B4) in endothelial-specific (VEC=VE-cadherin) PGC1α transgenic mice (VEC-tTA x TRE-PGC1α), n=5/group. Error bars SEM, *p < 0.05.
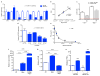
a, Gene expression for de novo NAD biosynthetic pathway in renal tubular cells 48 h after control vs. PGC1α knockdown (n=3/group). The gene expression set corresponds to the eight transcripts whose abundance was measured in kidney homogenates in Figure 3. P=0.0001 by two-way ANOVA with Bonferroni-corrected comparisons as indicated. b, Correlation of renal Nam vs. renal NAD in mice treated with vehicle or different doses of Nam (100–400 mg/kg IP × 1). Arbitrary units on X- and Y-axes. c, Renal β-OHB concentrations in kidneys of mice following vehicle (Veh) vs. Nam treatment (400 mg/kg IP × 4 d) with and without IRI 24 h prior to tissue collection (n=5/group). P-value by two-way ANOVA. Dashed line indicates normal circulating concentration of β-OHB. d, Dosing for siRNA against HCAR2 in renal tubular cells. e, Dose-inhibition curve in renal tubular cells for PGE2 release following 24 h of mepenzolate bromide at the indicated concentrations (n=3 replicates per concentration).– f,g Intracellular Nam and secreted β-OHB for renal tubular cells following treatment with Nam (1 μM for 24 h) with or without pre-treatment with the NAMPT inhibitor FK866 (10 nM, n=6/group). h, PGE2 in conditioned media of renal tubular cells after control vs. PGC1α knockdown and with and without exogenous β-OHB application (+, 5 mM, n=6/group, p values vs. control group). Error bars SEM, *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
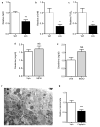
a–c, Relative renal NAD, β-OHB, and PGE2 concentrations in WT littermates vs. PGC1α−/− (KO) mice (n=6/group). d, Serum creatinine in genetic control mice for iNephPGC1α 24 h after IRI with vehicle vs. mepenzolate (MPN, 10 mg/kg IP) treatment (n=5/group). e, Serum creatinine in genetic control mice for iNephPGC1α 24 h after IRI with vehicle vs. indomethacin (INDO, 10 mg/kg IP) treatment (n=6/group). f, Transmission EM with cytochrome c oxidase enzyme histochemistry of proximal tubular cell 24 h following cisplatin exposure (25 mg/kg IP) demonstrating mitochondrial injury. Scale bar 500 nm. g, Relative renal Nam concentrations following cisplatin as in f. Error bars SEM, *p<0.05, **p<0.01, ***p<0.001.

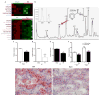
a–d, Low (a,b) and high-power (c,d) photomicrographs of PGC1α immunoreactivity (brown) in wildtype littermates (WT) and PGC1α−/− (KO) kidneys. Scale bars 100 and 50 μm. e,f Representative results of peptide competition attenuating PGC1α immunoreactivity against human kidney (n=4) as described in Methods. g, Representative immunostaining (brown) for PGC1α in a renal biopsy with chronic kidney disease (CKD). Scale bar 50 μm. h, Results of scoring PGC1α immunostaining intensity (1=weakest, 4=strongest) in specimens with CKD by blinded operator. Each dot represents a unique specimen. Analyzed by Mann-Whitney.
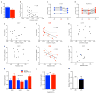
a, Serum creatinine in uninduced (−Dox) vs. induced (+Dox) iNephPGC1α mice (n = 8–10 mice per group). b, Comparison of serum creatinine with degree of renal PGC1α expression, p<0.05. c,d Serial serum creatinines in iNephPGC1α mice vs. controls before PGC1α induction (OFF), after 4 weeks of PGC1α induction (ON), and after 4 weeks of washout (OFF), *p<0.05 by repeated measures ANOVA. e–g, Comparison of serum creatinine at different time points with renal artery flow in iNephPGC1α mice from d, p<0.05 when correlation coefficient r = −0.65. h–j, Comparison of resistive index with renal artery flow volume in iNephPGC1α mice from d, p<0.05 when correlation coefficient r = −0.80. k, Relative renal expression of VEGF and nitric oxide synthases 1 and 3 (n=6/group). Analyzed by two-way ANOVA with Bonferroni corrections. l, Circulating thyroxine levels in iNephPGC1α mice with and without gene induction (n=5/group) to rule out Pax8-related thyrotoxicosis driving perfusion differences as previously described. m, Relative renal expression for VEGF in PGC1α−/− mice (KO) vs. WT littermates (n=6/group). Error bars SEM.
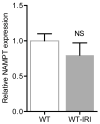
Relative renal expression for NAMPT in wildtype mice before and 24 h after IRI (n=6/group). Error bars SEM.
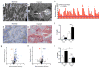
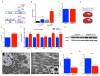
a, Pre-ischemic normal morphology and b, swollen mitochondria inside tubular cell 24h after ischemia-reperfusion injury (IRI). Scale bar 200nm. c, Renal di-/tri-acylglycerols (DAGs, TAGs) 24h following sham or IRI (n=6/group). P-value by ANOVA. d,e, Oil Red O (pink) for fat in normal and post-ischemic kidneys, scale bar 20μm. f, serum creatinine wildtype (WT) vs PGC1α−/− (KO) mice (basal, n=7/group; post-ischemia, n=18/group). g,h, Volcano plots of kidney metabolites from KO vs. WT or IRI vs. sham (univariate p<0.05 for colored dots, n=6/group). i, Serum creatinine in post-ischemic WT vs. KO mice treated with vehicle (Veh, n=5) vs. Nam (n=9). Error bars SEM, *p<0.05, **p<0.01.

a,b, Renal cytochrome c oxidase activity (brown), scale bar 500μm. c, Renal PGC1α and cytochrome c oxidase subunit IV. d, Survival curve following IRI (n=14 control; 24 iNephPGC1α). Dashed line for sham-operated mice (n=10). e, Serial serum creatinines from mice in d analyzed by ANOVA. f,g, Renal artery pulse wave and color Doppler 24h after IRI representative of 6/group. h–k, Tubular injury in cortex and outer stripe of outer medulla (OSOM) 24h after IRI representative of 8/group. Scale bar 100μm. l–o, Oil red O (pink) for fat in iNephPGC1α mice and controls 24h after IRI representative of 8/group. Scale bars 200μm (upper) and 50μm (lower). p, Renal Di-/tri-acylglycerols (DAGs, TAGs) in post-ischemic iNephPGC1α mice vs. controls (n=6/group). q, Relative renal Nam 24h after IRI (n=6/group). Error bars SEM, *p<0.05.
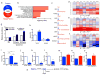
a, Renal RNA sequencing 24h after IRI or sham operation in controls vs. iNephPGC1α mice with enumerated transcripts. b, Pathway analysis of 1160 transcripts unique to post-ischemic iNephPGC1α mice graphed by −log10[Benjamini-Hochberg-corrected p-value]. Dashed line at p<0.05. c, de novo NAD biosynthetic pathway adapted from KEGG (
www.genome.jp/kegg). Trp=tryptophan, Kyn=kynurenine, Am=amino, Na=nicotinate. d–f, Heatmaps (red=lower, blue=higher) of intrarenal expression for de novo pathway from KO vs. WT; 24h after sham vs. IRI; and iNephPGC1α vs. controls (n=6/group). P-values by ANOVA. g, Relative renal Nam and NAD 4h after indicated Nam dose. P-value by ANOVA. h, Conditioned-media-PGE2 of renal tubular cells after HCAR2 knockdown with and without HCAR2 stimulation (+,niacin 10mM, n=6/group). i, PGE2 from renal cells following Nam (1μM for 24h) with and without NAMPT inhibitor FK866 (10nM, n=6/group). j–l, Intracellular NAD, conditioned-media beta-hydroxybutyrate (β-OHB), and conditioned-media PGE2 in PGC1α knockdown cells (n=6/group). m–o, Relative renal NAD, β-OHB, and PGE2 in control vs. iNephPGC1α mice (n=6/group). *p<0.05, **p<0.01, ***p<0.001. p, Renal epithelial PGC1α coordinately upregulates de novo NAD biosynthesis, in the absence of which Nam is utilized through the NAMPT-salvage pathway to generate NAD. Consequently, β-OHB accumulates, which signals HCAR2 to induce PGE2. Error bars SEM.
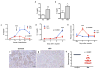
a, Serum creatinine in iNephPGC1α mice 24h after IRI with vehicle vs. mepenzolate (MPN, 10mg/kg IP) treatment (n=6/group). b, Serum creatinine in iNephPGC1α mice 24h after IRI with vehicle vs. indomethacin (INDO, 10mg/kg IP) treatment (n=6/group). c, Serial serum creatinines in mice receiving a single dose of Nam (400 mg/kg IP) 18h after the onset of reperfusion, i.e., with established AKI. Analyzed by ANOVA (n=5/group). d, Serial serum creatinines after cisplatin (25mg/kg IP administered on day 0) with or without Nam (400mg/kg IP on day −1 and day 0). Analyzed with Bonferroni-corrected ANOVA (n=5/group). e, Relative renal Nam from d. f,g, Representative immunostaining (brown) for PGC1α from control human kidney and a renal biopsy for AKI. Scale bars 50μm. h, PGC1α immunostaining intensity (1=weakest, 4=strongest). Each dot represents a unique specimen. Analyzed by Mann-Whitney. Error bars SEM, *p<0.05, **p<0.01, ****p<0.0001.
Comment in
-
Acute kidney injury: Improved fuel metabolism protects against AKI.
Allison SJ. Allison SJ. Nat Rev Nephrol. 2016 May;12(5):255. doi: 10.1038/nrneph.2016.45. Epub 2016 Mar 30. Nat Rev Nephrol. 2016. PMID: 27026351 No abstract available.
Similar articles
-
Collier JB, Schnellmann RG. Collier JB, et al. Cell Mol Life Sci. 2020 Sep;77(18):3643-3655. doi: 10.1007/s00018-019-03391-z. Epub 2019 Dec 23. Cell Mol Life Sci. 2020. PMID: 31873757 Free PMC article.
-
Yang SJ, Choi JM, Kim L, Park SE, Rhee EJ, Lee WY, Oh KW, Park SW, Park CY. Yang SJ, et al. J Nutr Biochem. 2014 Jan;25(1):66-72. doi: 10.1016/j.jnutbio.2013.09.004. Epub 2013 Oct 10. J Nutr Biochem. 2014. PMID: 24314867
-
Faivre A, Katsyuba E, Verissimo T, Lindenmeyer M, Rajaram RD, Naesens M, Heckenmeyer C, Mottis A, Feraille E, Cippà P, Cohen C, Longchamp A, Allagnat F, Rutkowski JM, Legouis D, Auwerx J, de Seigneux S. Faivre A, et al. Nephrol Dial Transplant. 2021 Jan 1;36(1):60-68. doi: 10.1093/ndt/gfaa124. Nephrol Dial Transplant. 2021. PMID: 33099633
-
PPARγ-Coactivator-1α, Nicotinamide Adenine Dinucleotide and Renal Stress Resistance.
Poyan Mehr A, Parikh SM. Poyan Mehr A, et al. Nephron. 2017;137(4):253-255. doi: 10.1159/000471895. Epub 2017 Jun 8. Nephron. 2017. PMID: 28591759 Free PMC article. Review.
-
Parikh SM. Parikh SM. Nephron. 2019;143(3):184-187. doi: 10.1159/000500168. Epub 2019 May 3. Nephron. 2019. PMID: 31055583 Free PMC article. Review.
Cited by
-
Standage SW, Xu S, Brown L, Ma Q, Koterba A, Lahni P, Devarajan P, Kennedy MA. Standage SW, et al. Am J Physiol Renal Physiol. 2021 May 1;320(5):F984-F1000. doi: 10.1152/ajprenal.00582.2020. Epub 2021 Apr 12. Am J Physiol Renal Physiol. 2021. PMID: 33843271 Free PMC article.
-
Zsengellér ZK, Rosen S. Zsengellér ZK, et al. J Histochem Cytochem. 2016 Sep;64(9):546-55. doi: 10.1369/0022155416660291. J Histochem Cytochem. 2016. PMID: 27578326 Free PMC article.
-
Jotwani V, Yang SY, Thiessen-Philbrook H, Parikh CR, Katz R, Tranah GJ, Ix JH, Cummings S, Waikar SS, Shlipak MG, Sarnak MJ, Parikh SM, Arking DE. Jotwani V, et al. Hum Genet. 2024 Feb;143(2):151-157. doi: 10.1007/s00439-023-02615-4. Epub 2024 Feb 13. Hum Genet. 2024. PMID: 38349571 Free PMC article.
-
Tatsumi S, Katai K, Kaneko I, Segawa H, Miyamoto KI. Tatsumi S, et al. Pflugers Arch. 2019 Jan;471(1):109-122. doi: 10.1007/s00424-018-2204-2. Epub 2018 Sep 14. Pflugers Arch. 2019. PMID: 30218374 Review.
-
Tubular Mitochondrial Dysfunction, Oxidative Stress, and Progression of Chronic Kidney Disease.
Fontecha-Barriuso M, Lopez-Diaz AM, Guerrero-Mauvecin J, Miguel V, Ramos AM, Sanchez-Niño MD, Ruiz-Ortega M, Ortiz A, Sanz AB. Fontecha-Barriuso M, et al. Antioxidants (Basel). 2022 Jul 12;11(7):1356. doi: 10.3390/antiox11071356. Antioxidants (Basel). 2022. PMID: 35883847 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous