A human microprotein that interacts with the mRNA decapping complex - PubMed
A human microprotein that interacts with the mRNA decapping complex
Nadia G D'Lima et al. Nat Chem Biol. 2017 Feb.
Abstract
Proteomic detection of non-annotated microproteins indicates the translation of hundreds of small open reading frames (smORFs) in human cells, but whether these microproteins are functional or not is unknown. Here, we report the discovery and characterization of a 7-kDa human microprotein we named non-annotated P-body dissociating polypeptide (NoBody). NoBody interacts with mRNA decapping proteins, which remove the 5' cap from mRNAs to promote 5'-to-3' decay. Decapping proteins participate in mRNA turnover and nonsense-mediated decay (NMD). NoBody localizes to mRNA-decay-associated RNA-protein granules called P-bodies. Modulation of NoBody levels reveals that its abundance is anticorrelated with cellular P-body numbers and alters the steady-state levels of a cellular NMD substrate. These results implicate NoBody as a novel component of the mRNA decapping complex and demonstrate potential functionality of a newly discovered microprotein.
Figures

(a) K562 and HEK293T cellular peptides were enriched and subjected to multidimensional LC-MS proteomics. Peptide mass spectra were searched against a custom protein database obtained from the 3-frame translation of RNA-Seq data from these cell lines. Annotated peptides were removed by BLAST search to afford a list of non-annotated peptides. This workflow led to the discovery of a tryptic peptide (underlined sequence) derived from a polypeptide translated from a sORF (black) in the LOC550643 RNA transcript (gray). The polypeptide is hereafter referred to as NoBody. (b) Transfection of an expression construct corresponding to the annotated full-length LOC550643 cDNA sequence (gray), with an epitope tag (red) at the C-terminus of the putative short ORF (black) into HEK293T cells resulted in expression of NoBody (red anti-FLAG immunofluorescence image superimposed on differential interference contrast (DIC) image). Scale bar, 20 µm. (c) ClustalW2 alignment of full-length NoBody polypeptide sequence from a variety of mammals. Amino acid identity is indicated by asterisks.

(a) Quantitative proteomics of FLAG-NoBody immunoprecipitates from HEK293T lysates identified putative NoBody interaction partners (N = 1) (see Supplementary Data 1 for total list). Red bars correspond to proteins with a role in mRNA decapping. (b) Confirmation of NoBody-interaction partners by anti-FLAG immunoprecipitation and Western blotting (N = 3). (c) Anti-c-myc immunoprecipitation of c-myc-EDC4 and c-myc-Dcp1A from HEK293T cells co-expressing FLAG-NoBody (cells expressing NoBody-FLAG alone as a negative control) followed by anti-FLAG immunoblotting demonstrates that FLAG-NoBody is enriched by EDC4. (d) HEK293T cells were transfected with either non-targeting siRNA, or siRNA targeting EDC4, Dcp1A, or Dcp2. 48 hours later, cells were lysed and recombinantly expressed and purified FLAG-NoBody was added. Anti-FLAG immunoprecipitation (IP) isolated NoBody-interacting complexes. (e) NoBody was labeled with the photo-cross-linker 4-(Maleimido)benzophenone (NoBody-BP) and irradiated at 365 nm in the presence or absence of purified FLAG-EDC4. In the presence of EDC4, a higher molecular weight cross-linked product is observed at a molecular weight that would correspond to a 1:1 complex between EDC4 and NoBody.

(a) In order to determine the region of the NoBody peptide responsible for interacting with EDC4, a series of 10-amino-acid deletions spanning the length of NoBody was prepared and fused to EGFP to equalize their expression. The constructs were co-transfected with c-myc-EDC4 in HEK293T cells, then lysates were subjected to anti-myc immunoprecipitation followed by anti-GFP Western blotting. 5% of each lysate and 25% of each immunoprecipitate was loaded in each lane. (b,c) The 20-amino-acid fragment of NoBody required for EDC4 co-precipitation (NoBody(22–41)) was fused to the N- and C-termini of EGFP to assay its sufficiency for interaction with EDC4. The NoBody(22–41)-EGFP fusions were co-transfected into HEK293T cells with c-myc-EDC4, then lysates subjected to anti-c-myc immunoprecipitation followed by anti-GFP Western blotting. A negative control (d) was performed with non-fused EGFP alone. 2–5 % of lysate and 40–50 % of each immunoprecipitate was loaded in each lane. (e) Alanine scanning mutagenesis of full-length FLAG-tagged NoBody peptide between NoBody amino acid residues 22 to 41 was performed to identify residues essential for the interaction with EDC4. These constructs were co-expressed with c-myc-EDC4 in HEK293T cells and subjected to anti-FLAG immunoprecipitation, followed by anti-c-myc Western blotting to assess EDC4 interaction.
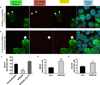
HEK293T cells were transfected with FLAG-NoBody via lentiviral infection at low viral titer. After fixation cells were stained with antibodies detecting a P-body marker (Dcp1A, red) and FLAG-NoBody (green), then counterstained with DAPI to visualize the nucleus (blue). In the merged image, NoBody/Dcp1A co-localization appears yellow. (a) In a minority of transfected cells (~10%), NoBody forms puncta that co-localize with P-bodies (starred). (b) In a majority of transfected cells (~90%), over-expression of NoBody leads to an absence of macroscopically detectable P-bodies (arrow). (c) HEK293T cells were transfected with FLAG-NoBody or the non-EDC4-interacting mutant FLAG-NoBodyΔ32–41. Endogenous P-bodies were visualized with anti-Dcp1A. Four fields of view, including the representative images, were used to quantitate average P-bodies per cell, representing >377 cells, in each average. Data represent mean values +/− S.E.M. Data represent mean values +/− S.E.M, and significance was evaluated with ANOVA (d–e) HEK293T cells were transfected with non- or NoBody-silencing siRNA, then fixed and endogenous P-bodies were detected using either anti-EDC4 (d) or anti-Dcp1A (e) immunofluorescence. For quantitation of P-bodies per cell, >6 fields of view were analyzed, totaling >400 cells for each measurement. Data represent mean values +/− S.E.M, and significance was evaluated with a two-tailed t-test. Scale bars, 10 µm.
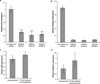
(a) Calu-6 cells were transfected with 2 independent siRNA sequences targeting NoBody for RNAi-mediated silencing, or with a non-targeting siRNA as a negative control. RNA was isolated 48 hours later, and levels of NoBody and p53 mRNA were determined by quantitative RT-PCR with reference to beta-actin. Data represent the mean of six biological replicates (**, p<0.01, ***, p<0.001). (b) NoBody silencing in Calu-6 was measured by quantitative RT-PCR with reference to beta-actin. (c) Calu-6 cells were lentivirally transfected for over-expression of the FLAG-NoBody coding sequence; untransfected cells served as a control. RNA was isolated 48 hours later, and levels of NoBody and p53 mRNA were determined by quantitative RT-PCR with reference to beta-actin. Data represent the mean of six biological replicates (*, p<0.05). (d) NoBody overexpression after lentiviral transfection in Calu-6 (b) was measured by quantitative RT-PCR with reference to beta-actin. Data represent mean values +/− standard deviation, and significance was evaluated with two-tailed t-test.
Similar articles
-
The NBDY Microprotein Regulates Cellular RNA Decapping.
Na Z, Luo Y, Schofield JA, Smelyansky S, Khitun A, Muthukumar S, Valkov E, Simon MD, Slavoff SA. Na Z, et al. Biochemistry. 2020 Oct 27;59(42):4131-4142. doi: 10.1021/acs.biochem.0c00672. Epub 2020 Oct 15. Biochemistry. 2020. PMID: 33059440 Free PMC article.
-
He F, Jacobson A. He F, et al. RNA. 2015 Sep;21(9):1633-47. doi: 10.1261/rna.052449.115. Epub 2015 Jul 16. RNA. 2015. PMID: 26184073 Free PMC article.
-
Structural and functional insights into eukaryotic mRNA decapping.
Ling SH, Qamra R, Song H. Ling SH, et al. Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):193-208. doi: 10.1002/wrna.44. Epub 2010 Sep 2. Wiley Interdiscip Rev RNA. 2011. PMID: 21957006 Review.
-
Mille viae in eukaryotic mRNA decapping.
Valkov E, Jonas S, Weichenrieder O. Valkov E, et al. Curr Opin Struct Biol. 2017 Dec;47:40-51. doi: 10.1016/j.sbi.2017.05.009. Epub 2017 Jun 4. Curr Opin Struct Biol. 2017. PMID: 28591671 Review.
Cited by
-
Webber CJ, Lei SE, Wolozin B. Webber CJ, et al. Prog Mol Biol Transl Sci. 2020;174:187-223. doi: 10.1016/bs.pmbts.2020.04.021. Epub 2020 May 12. Prog Mol Biol Transl Sci. 2020. PMID: 32828466 Free PMC article. Review.
-
RNA sequestration in P-bodies sustains myeloid leukaemia.
Kodali S, Proietti L, Valcarcel G, López-Rubio AV, Pessina P, Eder T, Shi J, Jen A, Lupión-Garcia N, Starner AC, Bartels MD, Cui Y, Sands CM, Planas-Riverola A, Martínez A, Velasco-Hernandez T, Tomás-Daza L, Alber B, Manhart G, Mayer IM, Kollmann K, Fatica A, Menendez P, Shishkova E, Rau RE, Javierre BM, Coon J, Chen Q, Van Nostrand EL, Sardina JL, Grebien F, Di Stefano B. Kodali S, et al. Nat Cell Biol. 2024 Oct;26(10):1745-1758. doi: 10.1038/s41556-024-01489-6. Epub 2024 Aug 21. Nat Cell Biol. 2024. PMID: 39169219
-
The NBDY Microprotein Regulates Cellular RNA Decapping.
Na Z, Luo Y, Schofield JA, Smelyansky S, Khitun A, Muthukumar S, Valkov E, Simon MD, Slavoff SA. Na Z, et al. Biochemistry. 2020 Oct 27;59(42):4131-4142. doi: 10.1021/acs.biochem.0c00672. Epub 2020 Oct 15. Biochemistry. 2020. PMID: 33059440 Free PMC article.
-
Kubatova N, Pyper DJ, Jonker HRA, Saxena K, Remmel L, Richter C, Brantl S, Evguenieva-Hackenberg E, Hess WR, Klug G, Marchfelder A, Soppa J, Streit W, Mayzel M, Orekhov VY, Fuxreiter M, Schmitz RA, Schwalbe H. Kubatova N, et al. Chembiochem. 2020 Apr 17;21(8):1178-1187. doi: 10.1002/cbic.201900677. Epub 2020 Jan 21. Chembiochem. 2020. PMID: 31705614 Free PMC article.
-
Westervelt N, Chadwick BP. Westervelt N, et al. Genome Biol Evol. 2018 Sep 1;10(9):2190-2204. doi: 10.1093/gbe/evy176. Genome Biol Evol. 2018. PMID: 30102341 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous