Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes - PubMed
- ️Mon Jan 01 2018
Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes
Jeffrey K Cornuault et al. Microbiome. 2018.
Abstract
Background: Viral metagenomic studies have suggested a role for bacteriophages in intestinal dysbiosis associated with several human diseases. However, interpretation of viral metagenomic studies is limited by the lack of knowledge of phages infecting major human gut commensal bacteria, such as Faecalibacterium prausnitzii, a bacterial symbiont repeatedly found depleted in inflammatory bowel disease (IBD) patients. In particular, no complete genomes of phages infecting F. prausnitzii are present in viral databases.
Methods: We identified 18 prophages in 15 genomes of F. prausnitzii, used comparative genomics to define eight phage clades, and annotated the genome of the type phage of each clade. For two of the phages, we studied prophage induction in vitro and in vivo in mice. Finally, we aligned reads from already published viral metagenomic data onto the newly identified phages.
Results: We show that each phage clade represents a novel viral genus and that a surprisingly large fraction of them (10 of the 18 phages) codes for a diversity-generating retroelement, which could contribute to their adaptation to the digestive tract environment. We obtained either experimental or in silico evidence of activity for at least one member of each genus. In addition, four of these phages are either significantly more prevalent or more abundant in stools of IBD patients than in those of healthy controls.
Conclusion: Since IBD patients generally have less F. prausnitzii in their microbiota than healthy controls, the higher prevalence or abundance of some of its phages may indicate that they are activated during disease. This in turn suggests that phages could trigger or aggravate F. prausnitzii depletion in patients. Our results show that prophage detection in sequenced strains of the microbiota can usefully complement viral metagenomic studies.
Keywords: Bacteriophages; Comparative genomics; Faecalibacterium prausnitzii; Inflammatory bowel disease; Prophages.
Conflict of interest statement
Ethics approval and consent to participate
All animal procedures were carried out according to the European Community Rules of Animal Care and with authorization 1234-2015101315238694 from French services.
Consent for publication
The manuscript does not contain any individual personal data in any form.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Whole genome dot plot of the 23 F. prausnitzii prophages and 7 homologous sequences retrieved from the nr/nt database define six clades and two singletons (Brigit and Oengus, last lanes). Braces group similar prophages, which correspond to the same phage species found in different bacterial genomes. Names beginning “VC” (underlined) correspond to metagenomic viral contigs, and names in italic correspond to prophages in non-F. prausnitzii bacterial species
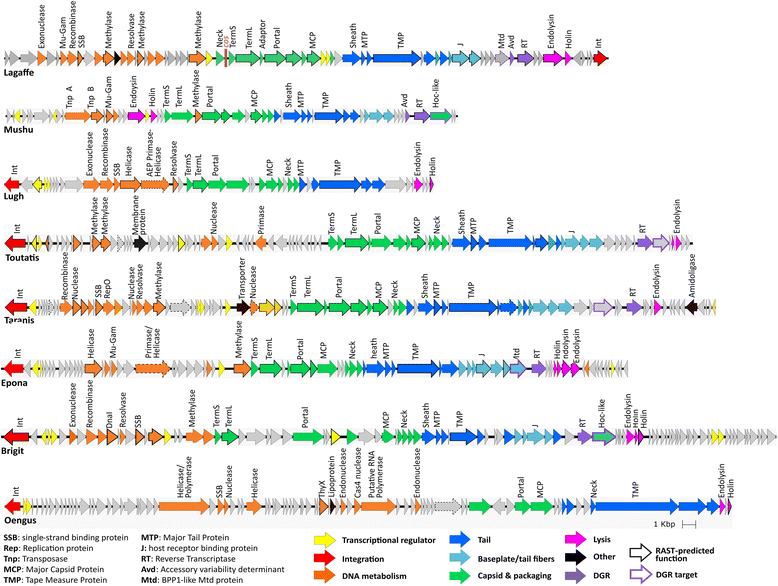
Genomes of each type phage. Genes whose product were predicted by RAST rather than by Phagonaute are indicated by a bold outline, and genes whose RAST-predicted function was changed in this study are indicated by a dashed outline
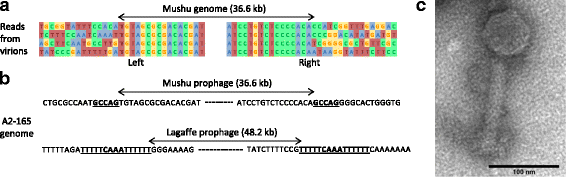
Mushu and Lagaffe are active phages. a Fragments of eight arbitrarily chosen reads of the two extremities of Mushu encapsidated DNA, showing the variable bacterial sequences on each side, typical of transpositional replication. b Direct repeats (underlined) on both sides of the prophages are present in the A2-165 genome. c Photograph of Mushu virion obtained by transmission electronic microcopy
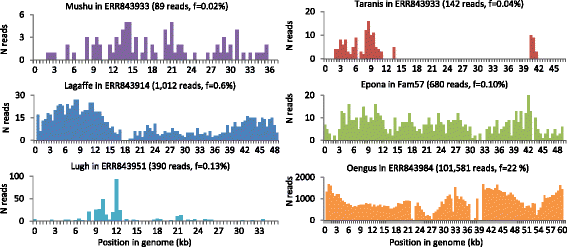
F. prausnitzii phages are present in intestinal viromes. Read coverage for six type phages in samples with the best abundance and coverage from the datasets of Norman et al. [18] and Reyes et al. [52]. Number of aligned reads (ordinate) is represented as a function of position in the phage genome (abscissa). Phage name, virome name, total number of aligned reads, and ratio of aligned to total reads are shown for each panel
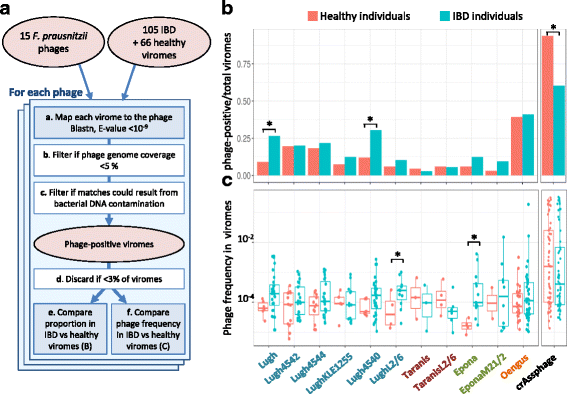
F. prausnitzii phages in healthy versus IBD-diseased individuals from the dataset of Norman et al. [18]. a Workflow of the procedure. a To infer the presence of a phage in a given sample, virome reads were aligned to the phage genome, and hits showing greater than 75% identity were retained. b If hits of a given sample were concentrated on less than 5% of the phage genome, the sample was not considered further for this phage. c The number of hits was compared to the maximal theoretical number of hits that could result from the measured bacterial DNA contamination. If the number of hits was significantly higher, the sample was classified as phage-positive. d If phage-positive samples represent less than 3% of total samples, this phage was not further studied. e The proportion of phage-positive viromes is compared to IBD versus healthy viromes. f In phage-positive viromes, phage frequency is compared to IBD versus healthy samples. b Fraction of phage-positive samples according to the health status of individuals. Three phages are significantly more prevalent in IBD-diseased than in healthy individuals. The highly prevalent crAssphage was included as a positive control in the analysis. c Phage abundance (read frequency) in positive samples according to the health status of individuals. Two phages, Epona and LughL2/6, are significantly more abundant in IBD patients than in healthy individuals
Similar articles
-
The Human Gut Phage Community and Its Implications for Health and Disease.
Manrique P, Dills M, Young MJ. Manrique P, et al. Viruses. 2017 Jun 8;9(6):141. doi: 10.3390/v9060141. Viruses. 2017. PMID: 28594392 Free PMC article. Review.
-
Eppinga H, Sperna Weiland CJ, Thio HB, van der Woude CJ, Nijsten TE, Peppelenbosch MP, Konstantinov SR. Eppinga H, et al. J Crohns Colitis. 2016 Sep;10(9):1067-75. doi: 10.1093/ecco-jcc/jjw070. Epub 2016 Mar 12. J Crohns Colitis. 2016. PMID: 26971052
-
Xu J, Liang R, Zhang W, Tian K, Li J, Chen X, Yu T, Chen Q. Xu J, et al. J Diabetes. 2020 Mar;12(3):224-236. doi: 10.1111/1753-0407.12986. Epub 2019 Oct 30. J Diabetes. 2020. PMID: 31503404 Free PMC article.
-
Fitzgerald CB, Shkoporov AN, Sutton TDS, Chaplin AV, Velayudhan V, Ross RP, Hill C. Fitzgerald CB, et al. BMC Genomics. 2018 Dec 14;19(1):931. doi: 10.1186/s12864-018-5313-6. BMC Genomics. 2018. PMID: 30547746 Free PMC article.
-
Action and function of Faecalibacterium prausnitzii in health and disease.
Ferreira-Halder CV, Faria AVS, Andrade SS. Ferreira-Halder CV, et al. Best Pract Res Clin Gastroenterol. 2017 Dec;31(6):643-648. doi: 10.1016/j.bpg.2017.09.011. Epub 2017 Sep 18. Best Pract Res Clin Gastroenterol. 2017. PMID: 29566907 Review.
Cited by
-
Thousands of previously unknown phages discovered in whole-community human gut metagenomes.
Benler S, Yutin N, Antipov D, Rayko M, Shmakov S, Gussow AB, Pevzner P, Koonin EV. Benler S, et al. Microbiome. 2021 Mar 29;9(1):78. doi: 10.1186/s40168-021-01017-w. Microbiome. 2021. PMID: 33781338 Free PMC article.
-
Gulyaeva A, Garmaeva S, Kurilshikov A, Vich Vila A, Riksen NP, Netea MG, Weersma RK, Fu J, Zhernakova A. Gulyaeva A, et al. Viruses. 2022 Oct 20;14(10):2305. doi: 10.3390/v14102305. Viruses. 2022. PMID: 36298860 Free PMC article.
-
Phages and their potential to modulate the microbiome and immunity.
Federici S, Nobs SP, Elinav E. Federici S, et al. Cell Mol Immunol. 2021 Apr;18(4):889-904. doi: 10.1038/s41423-020-00532-4. Epub 2020 Sep 8. Cell Mol Immunol. 2021. PMID: 32901128 Free PMC article. Review.
-
Dietary D-xylose promotes intestinal health by inducing phage production in Escherichia coli.
Hu J, Wu Y, Kang L, Liu Y, Ye H, Wang R, Zhao J, Zhang G, Li X, Wang J, Han D. Hu J, et al. NPJ Biofilms Microbiomes. 2023 Oct 11;9(1):79. doi: 10.1038/s41522-023-00445-w. NPJ Biofilms Microbiomes. 2023. PMID: 37821428 Free PMC article.
-
Morphological and Genetic Characterization of Eggerthella lenta Bacteriophage PMBT5.
Sprotte S, Rasmussen TS, Cho GS, Brinks E, Lametsch R, Neve H, Vogensen FK, Nielsen DS, Franz CMAP. Sprotte S, et al. Viruses. 2022 Jul 22;14(8):1598. doi: 10.3390/v14081598. Viruses. 2022. PMID: 35893664 Free PMC article.
References
-
- Quevrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermudez-Humaran LG, Pigneur B, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65(3):415–425. doi: 10.1136/gutjnl-2014-307649. - DOI - PMC - PubMed
-
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources