Emerging Patterns in HIV-1 gp120 Variable Domains in Anatomical Tissues in the Absence of a Plasma Viral Load - PubMed
Emerging Patterns in HIV-1 gp120 Variable Domains in Anatomical Tissues in the Absence of a Plasma Viral Load
Susanna L Lamers et al. AIDS Res Hum Retroviruses. 2019 Jun.
Abstract
The HIV envelope protein contains five hypervariable domains (V1-V5) that are fundamental for cell entry. We contrasted modifications in the variable domains derived from a panel of 24 tissues from 7 subjects with no measurable plasma viral load (NPVL) to variable domains from 76 tissues from 15 subjects who had a detectable plasma viral load (PVL) at death. NPVL subject's V1 and V2 domains were usually highly length variable, whereas length variation in PVL sequences was more conserved. Longer V1s contained more charged residues, whereas longer V2s were more glycosylated. Structural analysis demonstrated V1/V2 charge, and N-site additions/subtractions were localized to the CD4 binding pocket. Diversified envelopes in tissues during therapy may represent a mechanism for HIV persistence in tissues, as binding pocket complexity is associated with HIV that may escape neutralization, whereas shorter envelopes are associated with increased infectivity. Further analysis of tissue-derived envelope sequences may enable better understanding of potential immunological approaches targeting the persistent HIV reservoir.
Keywords: HIV envelope; envelope diversity; tissue-based reservoirs.
Conflict of interest statement
No competing financial interests exist.
Figures
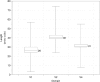
Length distribution from publicly held V1, V2, and V4 domains at the Los Alamos HIV database. The box spans the interquartile range, with the top and bottom representing the upper and lower quartiles of the data. The median value is shown and marked by a vertical line inside the box. The whiskers represent the highest and lowest observations.
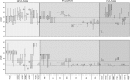
Env V1 (top) and V2 (bottom) length variation in individual anatomical tissues. Domain length is on y-axis, tissues sampled are on x-axis. NPVL = subjects who died with no detectable viral load. PVL = subjects who died with AIDS or a detectable viral load. Open circles in NPVL plots represent tissues where only one sequence was obtained. Dashed horizontal lines represent the upper and lower quartiles of the corresponding Los Alamos variable domains. SGS, single-genome sequencing; bPCR, bulk polymerase chain reaction.
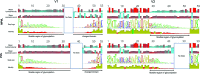
V1 and V2 amino acid variation in NPVL and PVL sequence alignments. Only nonidentical sequences were used in the alignment (NPVL = 127, PVL = 347 for V1; NPVL = 146, PVL = 399 for V2). Four panels are shown for each alignment: mean hydrophobicity is the hydrophobicity at each residue averaged over all sequences. Mean isoelectric point refers to the pH, at which a molecule carries no net electrical charge. “SeqLogo” displays amino acid, where the height of the amino acid is proportional to its frequency at each position in the alignment. “Identity” graphically displays the conservation of amino acids across all sequences for every position, where the height of the bar indicates the number of identical amino acids at each position; a green bar indicates that the residue at the position is the same across all sequences; yellow indicates less than complete identity; red indicates very low identity for a given position. Length flexible regions and charged regions are indicated. Color images are available online.

Structural variation in select long and short V1–V2 sequences. (A) Shows an alignment representative of a long and short V1–V2 sequence used for structural model development. Each sequence is identified by its GenBank accession number. The location of the V1 and V2 domains is shown on top of the alignment with a black bar. Amino acids are colored as follows: orange = N-sites, red = positively charged residues (K and R), blue = negatively charged residues (D, and E). To the right of the alignment, the total number of positively charged residues, negatively charged residues, net charge and number of glycosylation motifs for the entire V1V2 domain is noted. (B–F) 3D structural models developed using a protein threading technique for the sequences in (A). In (B), we show both models aligned and in cartoon format, with the long sequence colored green, and the short sequence colored cyan; the CD4 binding pocket is noted. (C) Contains the same aligned models as in (B), but here, we colored V1 red and V2 green. (D) Again, shows the same protein alignment as in (B), but here, we colored the positively and negatively charged residues on each structure using the same coloring scheme used in the alignment; any resides that are not charged or part of a N-site are colored green for the long sequence, and cyan for the short sequence. In (E), we have shown the same view as in (B–D); however, here, we have separated the long (top panel) from the short sequence (bottom panel) and the model is shown in “surface” format, with charged residues and N-sites colored. Note the dramatic differences in depth of the binding pocket between long and short V1–V2 sequences. In (F), we have rotated the models of (E) 90° to view the top of the structure and the inside of pocket that CD4 must traverse to successfully bind V3. The yellow lines in V1 contrasts the charged residues (blue and red) that line the inside of the pocket in long (top) and short (bottom) structures; whereas the yellow line in V2 highlights the glycosylation sites (orange) in V2. Note that in the long structure, 14 charged residues in V1 line the pocket and 3 N-sites completely circle the V2 arm of the pocket, whereas in the short structure, only six charged residues and one glycosylation site are present in the pocket. Color images are available online.

Structural variation in select long and short V1–V5 sequences. Note that for contrast, we used different sequences than used in Figure 4. In (A), we show the aligned sequences used to develop structures in (B, C), which are colored similarly to Figure 4. In (B), on the top left, we show a side view of the long structure. V1–V2 and V4–V5 are shown in cartoon format, and we highlight the V3 domain, which rests below V1/V2 in “sphere” format. On the right, V1–V2 is shown in a “surface” format to demonstrate the complete complex that CD4 must traverse for binding to occur. Below, we have rotated the model 90 degrees to show the top view of the binding pocket, with yellow lines highlighting the charged resides in V1 and V2. (C) Shows the short structure and the same views as in (B). Note the accumulation of charges (blue and red) in the pocket of the long V1 structure, compared with the short structure; also note the accumulation of glycosylation sites (orange) in the long V2 structure, similar to those observed in Figure 4. The functional progression of CD4 as well as coreceptor binding requires a movement of V1V2 to an “open” state allowing for exposure of V3; an extensive description of this process was recently published. Color images are available online.
Similar articles
-
Foda M, Harada S, Maeda Y. Foda M, et al. Microbiol Immunol. 2001;45(7):521-30. doi: 10.1111/j.1348-0421.2001.tb02653.x. Microbiol Immunol. 2001. PMID: 11529558
-
Brumme ZL, Dong WW, Yip B, Wynhoven B, Hoffman NG, Swanstrom R, Jensen MA, Mullins JI, Hogg RS, Montaner JS, Harrigan PR. Brumme ZL, et al. AIDS. 2004 Mar 5;18(4):F1-9. doi: 10.1097/00002030-200403050-00001. AIDS. 2004. PMID: 15090786
-
[Role of the HIV-1 gp120 V1/V2 domains in the induction of neutralizing antibodies].
Granados-González V, Piedrahita LD, Martínez M, Genin C, Riffard S, Urcuqui-Inchima S. Granados-González V, et al. Enferm Infecc Microbiol Clin. 2009 Nov;27(9):523-30. doi: 10.1016/j.eimc.2008.02.010. Epub 2009 May 1. Enferm Infecc Microbiol Clin. 2009. PMID: 19409660 Review. Spanish.
-
HIV-1 Reservoirs During Suppressive Therapy.
Barton K, Winckelmann A, Palmer S. Barton K, et al. Trends Microbiol. 2016 May;24(5):345-355. doi: 10.1016/j.tim.2016.01.006. Epub 2016 Feb 12. Trends Microbiol. 2016. PMID: 26875617 Free PMC article. Review.
Cited by
-
Silver S, Schmelz M. Silver S, et al. AIDS Res Ther. 2023 Aug 28;20(1):61. doi: 10.1186/s12981-023-00558-4. AIDS Res Ther. 2023. PMID: 37641153 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials