The role and mechanisms of DNA methylation in the oocyte - PubMed
- ️Tue Jan 01 2019
Review
The role and mechanisms of DNA methylation in the oocyte
Gintarė Sendžikaitė et al. Essays Biochem. 2019.
Abstract
Epigenetic information in the mammalian oocyte has the potential to be transmitted to the next generation and influence gene expression; this occurs naturally in the case of imprinted genes. Therefore, it is important to understand how epigenetic information is patterned during oocyte development and growth. Here, we review the current state of knowledge of de novo DNA methylation mechanisms in the oocyte: how a distinctive gene-body methylation pattern is created, and the extent to which the DNA methylation machinery reads chromatin states. Recent epigenomic studies building on advances in ultra-low input chromatin profiling methods, coupled with genetic studies, have started to allow a detailed interrogation of the interplay between DNA methylation establishment and chromatin states; however, a full mechanistic description awaits.
Keywords: chromatin; imprinting; methylation; oocytes.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

(A) At day 5 post-partum (5dpp), the majority of oocytes are non-growing (NGOs). Oocytes gradually grow postnatally in mice, which is manifested by an increase in diameter. Representative sizes of oocyte at different dpps are shown. DNA metyltransferases DNMT3A, DNMT3B and DNMT3L are increasingly expressed from early growing oocytes, which coincides with the start of DNA methylation establishment. DNMT expression peaks in FGOs, when DNA methylation is completed at approximately 40% genome coverage. Ovulation initiates maturation of the FGO and transition to MII stage, when a polar body is formed (depicted in grey) and the MII oocyte remains arrested until fertilisation. Oocyte transcription is globally repressed at the MII stage, corresponding with decrease in DNMT3 transcript levels. DNA methylation is depicted by the black line, relative expression levels for DNMTA/DNMT3B and DNMT3L are in violet and magenta, respectively. Abbreviation: dpp, days post partum. (B) Schematic representation of DNA methylation and histone marks in the FGO. Unlike the methylome of somatic cells, which shows relatively high and even levels of DNA methylation across the genome, the oocyte methylome has distinct bimodal and clustered hyper- and hypo-methylation domains. In somatic cells, promoter CpG methylation status is linked to expression, where actively transcribed genes tend to have an unmethylated promoter, but in oocytes this relationship is more ambiguous. Hypermethylated domains in the oocyte are associated with actively transcribed genes and the H3K36me3 mark. Levels of H3K36me3 positively correlate with levels of gene expression at the locus. Notably, a subset of oocyte transcripts arise from oocyte-specific or LTR-driven promoters. Hypomethylated domains span transcriptionally inactive parts of the genome and only a small fraction of the oocyte methylome falls into a partially methylated domain category; these domains tend to overlap intergenic areas. H3K4me3 in the FGO has a non-canonical distribution, where it spreads from promoters, forming broad low to medium enrichment domains over hypo- and partially methylated domains, but not hypermethylated domains. Similarly, H3K27me3 has a non-canonical distribution over hypo- and partially methylated domains, some of which overlap H3K4me3, forming bivalent chromatin. Bivalent chromatin in the oocyte does not fully recapitulate bivalency found in embryonic tissues, as the enrichment of H3K27me3 is lower. Meanwhile, H3K9me2 covers approximately a quarter of oocyte genome, again, with exclusion of hypermethylated domains. Height of the curves indicates relative enrichment of histone marks. Abbreviation: LTR, long terminal repeat.
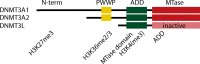
N-terminal domain of DNMT3A1 is required for localisation at bivalent chromatin shores in ES cells. Notably, this isoform is not expressed in the oocyte. The PWWP domain is poised to recognise H3K36me2/3, but this interaction has not been interrogated in the oocyte. The ADD domain forms a fold with the catalytic MTase domain to create an inactive allosteric conformation of DNMT3A. Recognition of unmethylated H3K4 tail alters this conformation and stimulates catalytic activity, while H3K4me3 repels the protein.
Similar articles
-
O'Doherty AM, O'Shea LC, Fair T. O'Doherty AM, et al. Biol Reprod. 2012 Mar 19;86(3):67. doi: 10.1095/biolreprod.111.094946. Print 2012 Mar. Biol Reprod. 2012. PMID: 22088914
-
Transcription and chromatin determinants of de novo DNA methylation timing in oocytes.
Gahurova L, Tomizawa SI, Smallwood SA, Stewart-Morgan KR, Saadeh H, Kim J, Andrews SR, Chen T, Kelsey G. Gahurova L, et al. Epigenetics Chromatin. 2017 May 12;10:25. doi: 10.1186/s13072-017-0133-5. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28507606 Free PMC article.
-
DNA Methyltransferases in Mammalian Oocytes.
Uysal F, Ozturk S. Uysal F, et al. Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review.
-
Lodde V, Modina SC, Franciosi F, Zuccari E, Tessaro I, Luciano AM. Lodde V, et al. Eur J Histochem. 2009 Dec 29;53(4):e24. doi: 10.4081/ejh.2009.e24. Eur J Histochem. 2009. PMID: 22073356 Free PMC article.
-
DNA methylation establishment during oocyte growth: mechanisms and significance.
Tomizawa S, Nowacka-Woszuk J, Kelsey G. Tomizawa S, et al. Int J Dev Biol. 2012;56(10-12):867-75. doi: 10.1387/ijdb.120152gk. Int J Dev Biol. 2012. PMID: 23417409 Review.
Cited by
-
A DNA Methylation Perspective on Infertility.
Shacfe G, Turko R, Syed HH, Masoud I, Tahmaz Y, Samhan LM, Alkattan K, Shafqat A, Yaqinuddin A. Shacfe G, et al. Genes (Basel). 2023 Nov 27;14(12):2132. doi: 10.3390/genes14122132. Genes (Basel). 2023. PMID: 38136954 Free PMC article. Review.
-
Molecular mechanisms underlying totipotency.
Ishiuchi T, Sakamoto M. Ishiuchi T, et al. Life Sci Alliance. 2023 Sep 4;6(11):e202302225. doi: 10.26508/lsa.202302225. Print 2023 Nov. Life Sci Alliance. 2023. PMID: 37666667 Free PMC article. Review.
-
Epigenetic reprogramming during the maternal-to-zygotic transition.
Chen Y, Wang L, Guo F, Dai X, Zhang X. Chen Y, et al. MedComm (2020). 2023 Aug 2;4(4):e331. doi: 10.1002/mco2.331. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37547174 Free PMC article. Review.
-
Lu YJ, Li Q, Chen LX, Tian T, Kang J, Hao YX, Zhou JS, Wang YY, Yan LY, Li R, Chang L, Qiao J. Lu YJ, et al. Hum Reprod Open. 2022 Dec 5;2023(1):hoac055. doi: 10.1093/hropen/hoac055. eCollection 2023. Hum Reprod Open. 2022. PMID: 36531663 Free PMC article.
-
Danzig J, Li D, Jan de Beur S, Levine MA. Danzig J, et al. J Clin Endocrinol Metab. 2021 Oct 21;106(11):e4603-e4620. doi: 10.1210/clinem/dgab460. J Clin Endocrinol Metab. 2021. PMID: 34157100 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources