A plasmid locus associated with Klebsiella clinical infections encodes a microbiome-dependent gut fitness factor - PubMed
- ️Fri Jan 01 2021
. 2021 Apr 30;17(4):e1009537.
doi: 10.1371/journal.ppat.1009537. eCollection 2021 Apr.
Christine M Bassis 3 , Srividya Ramakrishnan 4 , Robert Hein 3 , Sophia Mason 1 , Yehudit Bergman 5 , Nicole Sunshine 1 , Yunfan Fan 6 , Caitlyn L Holmes 1 2 , Winston Timp 6 7 8 , Michael C Schatz 4 9 10 , Vincent B Young 2 3 , Patricia J Simner 5 , Michael A Bachman 1 2
Affiliations
- PMID: 33930099
- PMCID: PMC8115787
- DOI: 10.1371/journal.ppat.1009537
A plasmid locus associated with Klebsiella clinical infections encodes a microbiome-dependent gut fitness factor
Jay Vornhagen et al. PLoS Pathog. 2021.
Abstract
Klebsiella pneumoniae (Kp) is an important cause of healthcare-associated infections, which increases patient morbidity, mortality, and hospitalization costs. Gut colonization by Kp is consistently associated with subsequent Kp disease, and patients are predominantly infected with their colonizing strain. Our previous comparative genomics study, between disease-causing and asymptomatically colonizing Kp isolates, identified a plasmid-encoded tellurite (TeO3-2)-resistance (ter) operon as strongly associated with infection. However, TeO3-2 is extremely rare and toxic to humans. Thus, we used a multidisciplinary approach to determine the biological link between ter and Kp infection. First, we used a genomic and bioinformatic approach to extensively characterize Kp plasmids encoding the ter locus. These plasmids displayed substantial variation in plasmid incompatibility type and gene content. Moreover, the ter operon was genetically independent of other plasmid-encoded virulence and antibiotic resistance loci, both in our original patient cohort and in a large set (n = 88) of publicly available ter operon-encoding Kp plasmids, indicating that the ter operon is likely playing a direct, but yet undescribed role in Kp disease. Next, we employed multiple mouse models of infection and colonization to show that 1) the ter operon is dispensable during bacteremia, 2) the ter operon enhances fitness in the gut, 3) this phenotype is dependent on the colony of origin of mice, and 4) antibiotic disruption of the gut microbiota eliminates the requirement for ter. Furthermore, using 16S rRNA gene sequencing, we show that the ter operon enhances Kp fitness in the gut in the presence of specific indigenous microbiota, including those predicted to produce short chain fatty acids. Finally, administration of exogenous short-chain fatty acids in our mouse model of colonization was sufficient to reduce fitness of a ter mutant. These findings indicate that the ter operon, strongly associated with human infection, encodes factors that resist stress induced by the indigenous gut microbiota during colonization. This work represents a substantial advancement in our molecular understanding of Kp pathogenesis and gut colonization, directly relevant to Kp disease in healthcare settings.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

(A) The ter locus is organized in two operons, a putative biosynthetic cluster and a TeO3-2 resistance cluster. These sections are found on opposite DNA strands and are encoded bidirectionally. The representative ter locus from the hvKp strain NTUH-K2044 is shown. NTUH-K2044 containing the empty vector pACYC184, the isogenic ΔterC mutant (clone Kp2259) containing an empty vector, the pTerC, or the pTerZ-F plasmid (B), and the E. coli K12 strain MG1655 with or without the pTerZ-F plasmid (C) were grown on LB or LB containing 10 or 100 μM K2TeO3-2 to visualize inhibition of growth (dilution series 100−10−7 of overnight culture). Two representative clones (labeled #1 and #2) of NTUH-K2044ΔterC containing the pTerC or the pTerZ-F plasmid and MG1655 containing the pTerZ-F plasmid are shown.

ter+ plasmids from Martin et al. mSystems, 2018 [10] (A-C) and reference strains from the NCBI database (D-F) were analyzed. (A,D) Relative frequencies of sequence types (ST) of Kp strains containing ter+ plasmids. HvKp sequence types previously associated with the ter operon are outlined in a dashed line. (B,E) Heat map of ter+ plasmid sequence similarity to genes known to influence infection and antibiotic resistance genes. Each row represents an individual plasmid in the order of S2 Table (Martin et al. mSystems, 2018 [10] index 1–14, NCBI reference strains index 15–102). The pK2044 hvKp plasmid is highlighted by the red box, and hypervirulent Kp sequence types (hvST) previously associated with the ter operon are indicated. (C,F) To determine if any neighboring gene was consistently associated with ter, the gene neighborhood of ter plasmids encoding the ter operon from Martin et al. mSystems, 2018 [10] was visualized (C) and the frequency of ORFs adjacent to the ter operon encoded on reference plasmids from the NCBI database was calculated (F).
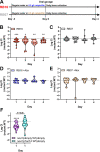
(A) Three days prior to inoculation, male and female C57BL6/J mice sourced from barriers RB16 and RB07 were treated with 0.5 g/L ampicillin or regular drinking water. (B-E) NTUH-K2044 and the isogenic ΔterC mutant (clone Kp2259) were mixed 1:1 and approximately 5x106 CFU were orally gavaged into mice (n = 9–18 per group). A fresh fecal pellet was collected daily from each animal, CFUs were enumerated, and log competitive indices (mutant:WT) were calculated (median and IQR displayed, *P < 0.05, **P < 0.005, ***P < 0.0005, one-sample t test compared to a hypothetical value of 0). (F) NTUH-K2044 and the isogenic ΔterC mutant containing an empty vector or the pTerZ-F plasmid were mixed 1:1 and approximately 5x106 CFU were orally gavaged into mice sourced from barrier RB16 (n = 14–16). A fresh fecal pellet was collected 24 hours after inoculation, CFUs were enumerated, and log competitive indices (mutant:WT) were calculated (F, median and IQR displayed, ****P < 0.00005, one-sample t test compared to a hypothetical value of 0 or Student’s t test). Each data point represents an individual animal.

Fecal pellets collected from male and female C57BL6/J mice sourced from barriers RB16 and RB07 (n = 9–20 mice per group) on the day of Kp inoculation were subjected to 16S rRNA sequencing. Pairwise community dissimilarity values between the fecal microbiota communities of barriers RB16 and RB07 with or without three days treatment with 0.5 g/L ampicillin were visualized by Principal coordinates analysis (A, AMOVA) and individually (B, **P < 0.005, ****P < 0.00005, one-way ANOVA followed by Tukey’s multiple comparisons post-hoc test). (C) Diversity of the fecal microbiota was summarized by inverse Simpson index (blue points: RB16+Abx, orange points: RB07+Abx, **P < 0.005, one-way ANOVA followed by Tukey’s multiple comparisons post-hoc test). LEfSe was used to determine if specific bacterial families (D) or OTUs (F) were differentially abundant between the fecal microbiota of RB16 and RB07 (D, LDA ≥ 3.5 and P < 0.05 are shown). Differential bacterial families (E) or OTUs (G) relative abundance values were plotted (E, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005, Student’s t test).
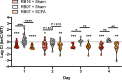
Seven days prior to inoculation, male and female C57BL6/J mice sourced from barriers RB16 and RB07 were treated with a SCFA cocktail or regular drinking water (sham). NTUH-K2044 and the isogenic ΔterC mutant (clone Kp2257) were mixed 1:1 and approximately 5x106 CFU were orally gavaged into mice (n = 19 per group). A fresh fecal pellet was collected daily from each animal, CFUs were enumerated, and log competitive indices (mutant:WT) were calculated (median and IQR displayed, *P < 0.05, **P < 0.005, ***P < 0.0005, one-sample t test compared to a hypothetical value of 0 or Holm-Sidak multiple-comparison test following one-way ANOVA).
Similar articles
-
The Klebsiella pneumoniae ter Operon Enhances Stress Tolerance.
Mason S, Vornhagen J, Smith SN, Mike LA, Mobley HLT, Bachman MA. Mason S, et al. Infect Immun. 2023 Feb 16;91(2):e0055922. doi: 10.1128/iai.00559-22. Epub 2023 Jan 18. Infect Immun. 2023. PMID: 36651775 Free PMC article.
-
Shimasaki T, Seekatz A, Bassis C, Rhee Y, Yelin RD, Fogg L, Dangana T, Cisneros EC, Weinstein RA, Okamoto K, Lolans K, Schoeny M, Lin MY, Moore NM, Young VB, Hayden MK; Centers for Disease Control and Prevention Epicenters Program. Shimasaki T, et al. Clin Infect Dis. 2019 May 30;68(12):2053-2059. doi: 10.1093/cid/ciy796. Clin Infect Dis. 2019. PMID: 30239622 Free PMC article.
-
Perez-Palacios P, Delgado-Valverde M, Gual-de-Torrella A, Oteo-Iglesias J, Pascual Á, Fernández-Cuenca F. Perez-Palacios P, et al. Appl Microbiol Biotechnol. 2021 Dec;105(24):9231-9242. doi: 10.1007/s00253-021-11684-2. Epub 2021 Nov 30. Appl Microbiol Biotechnol. 2021. PMID: 34846573
-
Boff L, de Sousa Duarte H, Kraychete GB, Taddeucci-Rocha G, Oliveira BD, Albano RM, D'Alincourt Carvalho-Assef AP, Superti SV, Martins IS, Picão RC. Boff L, et al. Infect Genet Evol. 2024 Jul;121:105598. doi: 10.1016/j.meegid.2024.105598. Epub 2024 Apr 21. Infect Genet Evol. 2024. PMID: 38653335
-
Host genetic control of gut microbiome composition.
Bubier JA, Chesler EJ, Weinstock GM. Bubier JA, et al. Mamm Genome. 2021 Aug;32(4):263-281. doi: 10.1007/s00335-021-09884-2. Epub 2021 Jun 22. Mamm Genome. 2021. PMID: 34159422 Free PMC article. Review.
Cited by
-
Pot M, Reynaud Y, Couvin D, Dereeper A, Ferdinand S, Bastian S, Foucan T, Pommier JD, Valette M, Talarmin A, Guyomard-Rabenirina S, Breurec S. Pot M, et al. Antibiotics (Basel). 2022 Oct 20;11(10):1443. doi: 10.3390/antibiotics11101443. Antibiotics (Basel). 2022. PMID: 36290101 Free PMC article.
-
Panchal J, Prajapati J, Dabhi M, Patel A, Patel S, Rawal R, Saraf M, Goswami D. Panchal J, et al. Mol Divers. 2024 Dec;28(6):3897-3918. doi: 10.1007/s11030-023-10785-6. Epub 2024 Jan 12. Mol Divers. 2024. PMID: 38212453
-
Wu X, Zhan F, Zhang J, Chen S, Yang B. Wu X, et al. Front Public Health. 2022 Aug 24;10:946370. doi: 10.3389/fpubh.2022.946370. eCollection 2022. Front Public Health. 2022. PMID: 36091562 Free PMC article.
-
Vávrová S, Grones J, Šoltys K, Celec P, Turňa J. Vávrová S, et al. Folia Microbiol (Praha). 2024 Apr;69(2):433-444. doi: 10.1007/s12223-024-01133-8. Epub 2024 Jan 23. Folia Microbiol (Praha). 2024. PMID: 38261148 Free PMC article.
-
Liang Z, Wang Y, Lai Y, Zhang J, Yin L, Yu X, Zhou Y, Li X, Song Y. Liang Z, et al. Front Cell Infect Microbiol. 2022 Nov 24;12:1050396. doi: 10.3389/fcimb.2022.1050396. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36506034 Free PMC article. Review.
References
-
- CDC. The Direct Medical Costs of Healthcare-Associated Infections in U.S. Hospitals and the Benefits of Prevention. 2009.
-
- O’Neill. Tackling Drug-Resistant Infections Globally: final report and recommendations 2016. [updated May 19]. Available from: https://amr-review.org.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases