Diffusive protein interactions in human versus bacterial cells - PubMed
- ️Wed Jan 01 2020
Diffusive protein interactions in human versus bacterial cells
Sarah Leeb et al. Curr Res Struct Biol. 2020.
Abstract
Random encounters between proteins in crowded cells are by no means passive, but found to be under selective control. This control enables proteome solubility, helps to optimise the diffusive search for interaction partners, and allows for adaptation to environmental extremes. Interestingly, the residues that modulate the encounters act mesoscopically through protein surface hydrophobicity and net charge, meaning that their detailed signatures vary across organisms with different intracellular constraints. To examine such variations, we use in-cell NMR relaxation to compare the diffusive behaviour of bacterial and human proteins in both human and Escherichia coli cytosols. We find that proteins that 'stick' in E. coli are generally less restricted in mammalian cells. Furthermore, the rotational diffusion in the mammalian cytosol is less sensitive to surface-charge mutations. This implies that, in terms of protein motions, the mammalian cytosol is more forgiving to surface alterations than E. coli cells. The cellular differences seem not linked to the proteome properties per se, but rather to a 6-fold difference in protein concentrations. Our results outline a scenario in which the tolerant cytosol of mammalian cells, found in long-lived multicellular organisms, provides an enlarged evolutionary playground, where random protein-surface mutations are less deleterious than in short-generational bacteria.
© 2020 The Author(s).
Conflict of interest statement
We declare no conflict of interest.
Figures
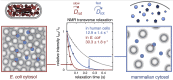
Properties of the E. coli and human cell cytosols, and their effect on protein stability. A. The cytosolic protein size distribution in E. coli (blue) and in human cells (red). The human proteome shows both a shift of the distribution peak towards larger protein size, as well as a significantly longer tail with larger proteins. Protein sequences of human (N = 5216) and E. coli cytosolic proteins (N = 1073) were collected from the Uniprot database (Consortium, 2018), in both cases the subset annotated as cytoplasmic was used for analysis. The distributions are fitted to a Γ-distribution. B. The surface net charge density of cytosolic proteins in E. coli (blue) and human cells (red). The charge density is somewhat higher in E. coli proteins, enabling stronger electrostatic repulsion. C. The temperature dependence of the folding free energy (ΔGN-D = −2.3RTlog [N]/[D], where [N] is the concentration of the folded and [D] of the unfolded state) of SOD1barrel shows the archetypical curvature of protein stability. SOD1barrel shows a significant destabilisation in A2780 cells (red) compared to in PBS-buffer (grey line). In the E. coli cytosol (blue line) the curve is shifted towards a higher melting temperature, while the maximum stability decreases significantly (Danielsson et al., 2015).

Improved in-cell NMR spectra in A2780 cells compared to E. coli. HMQC spectra of TTHApwt (blue frames), HAH1pwt (red frames) and SOD1barrel (green frames) in E. coli (dashed frames) and in live A2780 cells (solid frames). In E. coli, both HAH1pwt and SOD1barrel signals are severely broadened due to a high amount of interactions with the complex E. coli cytoplasm, i.e. the signals are barely visible in the HAH1pwt and SOD1barrel spectra even though the protein concentration is similar in all E. coli samples, as detected from the lysate signal intensity. In mammalian cells, all three proteins show significant improvement of the in-cell spectral properties, although still with varying line width. N.B. the contour levels differ in the E. coli dataset in order to visualise the broad low-intensity peaks of HAH1pwt and SOD1barrel. Right: the precited structures of TTHApwt (mutations templated on PDB id:
2ROE), HAH1pwt (mutations templated on PDB id:
1TL5) and the crystal structure of SOD1barrel (PDB id:
4BCZ) are shown. The calculated surface charge distribution is projected on the structure surfaces, with blue corresponding to positive charge density and red to negative, highlighting the differences in both charge distribution and charge clustering. Especially between the structural homologues TTHApwt and HAH1pwt severely broadened peaks. Even so, the line widths of the mammalian proteins narrow up in the cell lysates, showing that the Drot in the human cytosol is still reduced, albeit not as much as in E. coli (SI, Fig. S1). The results show thus that the mammalian cytosol affects the internalised proteins less than the E. coli cytosol, even to the extent that the protein-specific spectral characteristics are hard to distinguish at first sight.
First order quantification of the line broadening effect in A2780 cells. A-C. In-cell NMR spectra from A2780 cells in black overlaid by in-vitro spectra in dilute buffer in red, the frame colour code is as in Fig. 1. Inset in each spectrum is the average line width, determined from each well-separated cross peak. In D. the corresponding data for TTHApwt determined in E. coli is shown. E. The bars show the ratio between in-vitro and in-cell line widths.
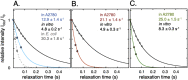
NMR relaxation data confirms the line broadening analysis. In-cell R2 relaxation rates in blue for TTHApwt (A.), red for HAH1pwt (B.) and in green SOD1barrel (C.), compared to the relaxation rates in dilute buffer (black solid lines). For all three proteins a significant increase in relaxation rate is observed, corresponding to a retardation of rotational diffusion. For TTHApwt, the R2 rate was determined also in E. coli, showing an even more pronounced retardation (grey dashed line in A.).
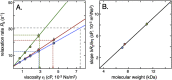
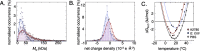
Standard curves for determination of apparent viscosity. A. Reference curves for how the average R2 rates of the three probe proteins (shown in Fig. 4) change with increasing microscopic viscosity (corresponding to pure ηint in Eq. (2)). Colour coding is as in Fig. 1, and error bars are the translated error from signal to noise in signal intensity determination. The slope δR2/δη is directly linked to the size of the protein, as highlighted in the correlation plot in B.

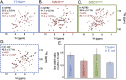
The apparent viscosity in A2780 human cells and in E. coli. A. The apparent viscosity, ηapp, shows a positive dependence on protein net charge, with increasing ηapp for positive protein surfaces. Colour codes are as in Fig. 1 and the dashed line is an exponential fit to the data points to guide the eye. The offset corresponds to water viscosity at infinite repulsion, while the exponentiality is adopted from the apparent exponential behaviour of the E. coli data. B. The same data as in the left panel compared to E. coli data (squares), N.B. the different axis scales. The large squares correspond to ηapp in E. coli for the three probe proteins, and the smaller faded squares to surface mutations, as determined from their in-cell NMR 1D-HMQC intensity. The blue triangle represents the apparent viscosity of TTHApwt in E. coli determined by the NMR relaxation rate R2, showing that the two methods are equivalent. Orange markers show the apparent viscosity for HAH1K57E. C. The observed ηapp in the two organisms correlate. The slope, however, is far from unity, indicating that the change in ηapp upon surface-charge mutation is 6 times stronger in E. coli.
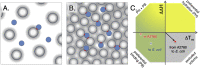
Snapshot representation of human and bacterial cytosol and thermodynamic classification of the effects from complex crowding solvents. A. A cartoon of the human cytosol with 60 mg/ml macromolecular concentration, here represented by ‘spherical’ proteins with the distribution of radii represented as soft edges. From relaxation data we can estimate that approximately 25% of the bacterial protein TTHApwt are involved in transient complexes at any given time point. B. The 6 times higher charge dependence on ηapp indicated 6 times higher macromolecular concentration in E. coli, and this results in that TTHApwt at any given time point is involved in a transient complex, and in more than 10% of the time it is involved in the formation of a transient trimer, with the slightly smaller proteins in the E. coli cytosol. C. The effects from altering the solvent can be classified from the effects on unfolding enthalpy (ΔH) and melting temperature (Tm), following the protocol by Ebbinghaus (Senske et al., 2014). The effects on SOD1barrel stability when comparing data from buffer to data from the A2780 cytosol is shown as a red ‘x’, and the corresponding effect when comparing to data from E. coli cytosol is marked as a blue ‘x’. Comparing human A2780 cell data to E. coli (red sphere) data results in a reduction in ΔH accompanied by an increase in Tm that can be classified as an excluded volume effect accompanied with increased transient binding, in line with an increased macromolecular concentration in the E. coli cytosol. The figure design is adapted from Senske et al. (Senske et al., 2014).
Similar articles
-
Physicochemical code for quinary protein interactions in Escherichia coli.
Mu X, Choi S, Lang L, Mowray D, Dokholyan NV, Danielsson J, Oliveberg M. Mu X, et al. Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):E4556-E4563. doi: 10.1073/pnas.1621227114. Epub 2017 May 23. Proc Natl Acad Sci U S A. 2017. PMID: 28536196 Free PMC article.
-
Diffusive intracellular interactions: On the role of protein net charge and functional adaptation.
Vallina Estrada E, Zhang N, Wennerström H, Danielsson J, Oliveberg M. Vallina Estrada E, et al. Curr Opin Struct Biol. 2023 Aug;81:102625. doi: 10.1016/j.sbi.2023.102625. Epub 2023 Jun 16. Curr Opin Struct Biol. 2023. PMID: 37331204 Review.
-
Physicochemical classification of organisms.
Vallina Estrada E, Oliveberg M. Vallina Estrada E, et al. Proc Natl Acad Sci U S A. 2022 May 10;119(19):e2122957119. doi: 10.1073/pnas.2122957119. Epub 2022 May 2. Proc Natl Acad Sci U S A. 2022. PMID: 35500111 Free PMC article.
-
Surface properties of the Vero cytotoxin-producing Escherichia coli O157:H7.
Sherman P, Soni R, Petric M, Karmali M. Sherman P, et al. Infect Immun. 1987 Aug;55(8):1824-9. doi: 10.1128/iai.55.8.1824-1829.1987. Infect Immun. 1987. PMID: 2886431 Free PMC article.
-
Nonadaptive processes in primate and human evolution.
Harris EE. Harris EE. Am J Phys Anthropol. 2010;143 Suppl 51:13-45. doi: 10.1002/ajpa.21439. Am J Phys Anthropol. 2010. PMID: 21086525 Review.
Cited by
-
Xiang L, Yan R, Chen K, Li W, Xu K. Xiang L, et al. Nano Lett. 2023 Mar 8;23(5):1711-1716. doi: 10.1021/acs.nanolett.2c04379. Epub 2023 Feb 20. Nano Lett. 2023. PMID: 36802676 Free PMC article.
-
Identifying the minimal sets of distance restraints for FRET-assisted protein structural modeling.
Liu Z, Grigas AT, Sumner J, Knab E, Davis CM, O'Hern CS. Liu Z, et al. ArXiv [Preprint]. 2024 Aug 19:arXiv:2405.07983v2. ArXiv. 2024. PMID: 38800659 Free PMC article. Updated. Preprint.
-
The intracellular environment affects protein-protein interactions.
Speer SL, Zheng W, Jiang X, Chu IT, Guseman AJ, Liu M, Pielak GJ, Li C. Speer SL, et al. Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2019918118. doi: 10.1073/pnas.2019918118. Proc Natl Acad Sci U S A. 2021. PMID: 33836588 Free PMC article.
-
Electron transfer in the respiratory chain at low salinity.
Lobez AP, Wu F, Di Trani JM, Rubinstein JL, Oliveberg M, Brzezinski P, Moe A. Lobez AP, et al. Nat Commun. 2024 Sep 19;15(1):8241. doi: 10.1038/s41467-024-52475-3. Nat Commun. 2024. PMID: 39300056 Free PMC article.
-
Choi AA, Xu K. Choi AA, et al. J Am Chem Soc. 2024 Apr 17;146(15):10973-10978. doi: 10.1021/jacs.4c02475. Epub 2024 Apr 4. J Am Chem Soc. 2024. PMID: 38576203
References
-
- Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. - PubMed
-
- Banci L., Bertini I., Calderone V., Della-Malva N., Felli I.C., Neri S., Pavelkova A., Rosato A. Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. Biochem. J. 2009;422:37–42. - PubMed
-
- Bas D.C., Rogers D.M., Jensen J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins. 2008;73:765–783. - PubMed
-
- Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3 - PubMed
LinkOut - more resources
Full Text Sources