Characterization of Escherichia coli harboring colibactin genes (clb) isolated from beef production and processing systems - PubMed
- ️Sat Jan 01 2022
Characterization of Escherichia coli harboring colibactin genes (clb) isolated from beef production and processing systems
Manita Guragain et al. Sci Rep. 2022.
Abstract
Certain strains of Escherichia coli possess and express the toxin colibactin (Clb) which induces host mutations identical to the signature mutations of colorectal cancer (CRC) that lead to tumorigenic lesions. Since cattle are a known reservoir of several Enterobacteriaceae including E. coli, this study screened for clb amongst E. coli isolated from colons of cattle-at-harvest (entering beef processing facility; n = 1430), across the beef processing continuum (feedlot to finished subprimal beef; n = 232), and in ground beef (n = 1074). Results demonstrated that clb+ E. coli were present in cattle and beef. Prevalence of clb+ E. coli from colonic contents of cattle and ground beef was 18.3% and 5.5%, respectively. clb+ E. coli were found susceptible to commonly used meat processing interventions. Whole genome sequencing of 54 bovine and beef clb+ isolates showed clb occurred in diverse genetic backgrounds, most frequently in phylogroup B1 (70.4%), MLST 1079 (42.6%), and serogroup O49 (40.7%).
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
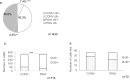
clb+ E. coli among cattle-at-harvest. (a) Percentage prevalence of clb+ E. coli among all generic E. coli isolated from cattle (b) Number of animals carrying clb+ E. coli. (c) Number of cattle lots carrying clb+ E. coli. Statistical significance was calculated by Fisher’s exact test. *, significant (p < 0.05); ***, extremely significant (p < 0.001). CONV, conventionally raised; RWA, Raised without antibiotics.

clb+ E. coli occurring in ground beef. (a) Percentage prevalence of clb+ E. coli in ground beef among all generic E. coli isolated; (b) Percentage of ground beef samples carrying clb+ E. coli. Statistical significance was calculated by Fisher’s exact test. *, significant (p < 0.05). CONV: ground beef produced from conventionally raised cattle; RWA: ground beef produced with label claim of “from beef raised without antibiotics”.
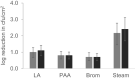
Beef flank inoculation assay. LA, 2% lactic acid (1 min). PAA, 200 ppm peroxyacetic acid (1 min). Brom, 300 ppm BoviBromine (1 min). Steam, flank surface temperature achieved 82 °C with dry steam (15 s). Statistical significance between clb+
E. coli and reference E. coli O157:H7 was calculated by Fisher’s exact test (p ≥ 0.05, not significant). Data presented represents average of 2 biological replicates, 12 technical replicates each on individual beef flanks.  clb+
E. coli,
clb+
E. coli,  E. coli O157:H7.
E. coli O157:H7.

Maximum likelihood phylogenetic tree of (a) 54 core genomes of clb+ E. coli isolated from beef production and processing continuum, and (b) 39 pks island sequences extracted from the contigs carrying full length pks island (clbA-clbS). Phylogenetic trees were constructed using IQ-tree and visualized with iTOL v6. Numbers on the outermost column represent multilocus sequence types (MLST).

Distribution of acquired antibiotic resistance genes in clb+ E. coli. Heat map was created and visualized using ggplot2 package of R. Column represents E. coli isolates and row represents acquired antibiotic resistance genes. *, Phylogroup B2; # Phylogroup A.

Distribution of acquired virulence genes in clb+ E. coli. Heat map was created and visualized using ggplot2 package of R. Column represents E. coli isolates and row represents acquired virulence genes. *, Phylogroup B2; # Phylogroup A.
Similar articles
-
Tariq H, Noreen Z, Ahmad A, Khan L, Ali M, Malik M, Javed A, Rasheed F, Fatima A, Kocagoz T, Sezerman U, Bokhari H. Tariq H, et al. PLoS One. 2022 Nov 11;17(11):e0262662. doi: 10.1371/journal.pone.0262662. eCollection 2022. PLoS One. 2022. PMID: 36367873 Free PMC article.
-
Yoshikawa Y, Tsunematsu Y, Matsuzaki N, Hirayama Y, Higashiguchi F, Sato M, Iwashita Y, Miyoshi N, Mutoh M, Ishikawa H, Sugimura H, Wakabayashi K, Watanabe K. Yoshikawa Y, et al. Jpn J Infect Dis. 2020 Nov 24;73(6):437-442. doi: 10.7883/yoken.JJID.2020.066. Epub 2020 May 29. Jpn J Infect Dis. 2020. PMID: 32475872
-
Hussein HS. Hussein HS. J Anim Sci. 2007 Mar;85(13 Suppl):E63-72. doi: 10.2527/jas.2006-421. Epub 2006 Oct 23. J Anim Sci. 2007. PMID: 17060419 Review.
-
Ekong PS, Sanderson MW, Cernicchiaro N. Ekong PS, et al. Prev Vet Med. 2015 Sep 1;121(1-2):74-85. doi: 10.1016/j.prevetmed.2015.06.019. Epub 2015 Jun 27. Prev Vet Med. 2015. PMID: 26153554 Review.
Cited by
-
The GEA pipeline for characterizing Escherichia coli and Salmonella genomes.
Dickey AM, Schmidt JW, Bono JL, Guragain M. Dickey AM, et al. Sci Rep. 2024 Jun 10;14(1):13257. doi: 10.1038/s41598-024-63832-z. Sci Rep. 2024. PMID: 38858528 Free PMC article.
References
-
- U.S Cancer Statistics Working group. U.S. Cancer statistics data visualizations tool, baed on 2020 submission data (1998–2018) (2021). www.cdc.gov/cancer/dataviz
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical