Genetic Structure of Racing Pigeons (Columba livia) Kept in Poland Based on Microsatellite Markers - PubMed
- ️Sat Jan 01 2022
Genetic Structure of Racing Pigeons (Columba livia) Kept in Poland Based on Microsatellite Markers
Angelika Podbielska et al. Genes (Basel). 2022.
Abstract
Pigeons played a major role in communication before the invention of the telephone and the telegraph, as well as in wars, where they were used to carry information and orders over long distances. Currently, numerous sports competitions and races are held with their participation, and their breeding is demanding not only for breeders, but also for the birds themselves. Therefore, an analysis of the genetic structure of racing pigeons kept in Poland was undertaken on the basis of 16 microsatellite markers, as well as the evaluation of the microsatellite panel recommended by ISAG. For this purpose, Bayesian clustering, a dendrogram, and Principal Coordinate Analysis were conducted. In addition, statistical analysis was performed. Based on this research, it was observed that racing pigeons are genetically mixed, regardless of their place of origin. Moreover, genetic diversity was estimated at a relatively satisfactory level (Ho = 0.623, He = 0.684), and no alarmingly high inbreeding coefficient was observed (F = 0.088). Moreover, it was found that the panel recommended by ISAG can be successfully used in Poland for individual identification and parentage testing (PIC = 0.639, CE-1P = 0.9987233, CE-2P = 0.9999872, CE-PP = 0.99999999).
Keywords: gene flow; genetic diversity; homing pigeons; individual identification; microsatellite markers; parentage testing; population structure; racing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
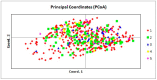
Principal coordinate analysis (PCoA) of all pigeons. 1—PL; 2—BE; 3—DV; 4—SK; 5—NL. Percentage of variation explained by the first 3 axes: 3.34, 2.89, and 2.45.

Dendrogram of genetic distance between all individuals with the UPGMA algorithm. Red—PL; green—BE; blue—DV; yellow—SK; pink—NL.
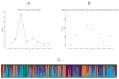
(A) Delta K values obtained with STRUCTURE HARVESTER. (B) Rate of change in the likelihood distribution (mean) obtained with STRUCTURE HARVESTER. (C) STRUCTURE software clustering at K = 4 on a dataset containing 519 individuals.
Similar articles
-
de Groot M, van Haeringen WA. de Groot M, et al. Anim Genet. 2017 Aug;48(4):431-435. doi: 10.1111/age.12555. Epub 2017 Apr 27. Anim Genet. 2017. PMID: 28449233
-
Radko A, Podbielska A. Radko A, et al. Genes (Basel). 2021 Mar 26;12(4):485. doi: 10.3390/genes12040485. Genes (Basel). 2021. PMID: 33810589 Free PMC article.
-
Radko A, Smołucha G, Koseniuk A. Radko A, et al. Genes (Basel). 2021 Apr 19;12(4):595. doi: 10.3390/genes12040595. Genes (Basel). 2021. PMID: 33921663 Free PMC article.
-
Marlier D. Marlier D. Vet Sci. 2022 Jan 22;9(2):42. doi: 10.3390/vetsci9020042. Vet Sci. 2022. PMID: 35202294 Free PMC article. Review.
-
Santos HM, Tsai CY, Catulin GEM, Trangia KCG, Tayo LL, Liu HJ, Chuang KP. Santos HM, et al. Vet Microbiol. 2020 Aug;247:108779. doi: 10.1016/j.vetmic.2020.108779. Epub 2020 Jun 28. Vet Microbiol. 2020. PMID: 32768225 Review.
References
-
- Ramadan S., Abe H., Hayano A., Yamaura J., Onoda T., Miyake T., Inoue-Murayama M. Analysis of genetic diversity of Egyptian pigeon breeds. J. Poult. Sci. 2011;48:79–84. doi: 10.2141/jpsa.010109. - DOI
-
- Od Dzikiego Gołębia Skalnego do ras Współcześnie Hodowanych—PDF Free Download. [(accessed on 21 April 2022)]. Available online: http://docplayer.pl/59099216-Od-dzikiego-golebia-skalnego-do-ras-wspolcz....
Publication types
MeSH terms
Grants and funding
This research was funded by the National Research Institute of Animal Production.
LinkOut - more resources
Full Text Sources
Miscellaneous