QTG-Miner aids rapid dissection of the genetic base of tassel branch number in maize - PubMed
- ️Sun Jan 01 2023
. 2023 Aug 26;14(1):5232.
doi: 10.1038/s41467-023-41022-1.
Juan Li # 1 2 , Linqian Han 1 2 , Chengyong Liang 1 2 , Jiaxin Li 1 2 , Xiaoyang Shang 1 2 , Xinxin Miao 1 2 , Zi Luo 1 2 , Wanchao Zhu 1 2 , Zhao Li 1 2 , Tianhuan Li 1 2 , Yongwen Qi 3 , Huihui Li 4 , Xiaoduo Lu 5 , Lin Li 6 7
Affiliations
- PMID: 37633966
- PMCID: PMC10460418
- DOI: 10.1038/s41467-023-41022-1
QTG-Miner aids rapid dissection of the genetic base of tassel branch number in maize
Xi Wang et al. Nat Commun. 2023.
Abstract
Genetic dissection of agronomic traits is important for crop improvement and global food security. Phenotypic variation of tassel branch number (TBN), a major breeding target, is controlled by many quantitative trait loci (QTLs). The lack of large-scale QTL cloning methodology constrains the systematic dissection of TBN, which hinders modern maize breeding. Here, we devise QTG-Miner, a multi-omics data-based technique for large-scale and rapid cloning of quantitative trait genes (QTGs) in maize. Using QTG-Miner, we clone and verify seven genes underlying seven TBN QTLs. Compared to conventional methods, QTG-Miner performs well for both major- and minor-effect TBN QTLs. Selection analysis indicates that a substantial number of genes and network modules have been subjected to selection during maize improvement. Selection signatures are significantly enriched in multiple biological pathways between female heterotic groups and male heterotic groups. In summary, QTG-Miner provides a large-scale approach for rapid cloning of QTGs in crops and dissects the genetic base of TBN for further maize breeding.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

QTG-Miner integrates three procedures: 1) primary QTL mapping, 2) screening and sequencing on single QTL segregating material, and 3) candidate gene mining by QTG-Miner. In 1), mapping population types include but are not limited to F2, DHs, RILs, and NILs. In 2), single QTL-paired materials are screened, planted, and sampled for RNA-seq. Identified DEGs and sequence variants are considered as proportional weights during candidate gene prediction. In 3), SD and ML algorithms are integrated into QTG-Miner. Using SD and ML algorithms, candidate genes underlying the target QTL can be uncovered and subsequently verified by EMS mutagenesis or CRISPR/Cas9-mediated editing.
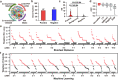
a Multi-omics network map used by QTG-Miner. ChIA-PET, chromatin interaction analysis by paired-end tag sequencing. PPI, protein-protein interaction. b Positive and negative gene datasets used for the SD and ML algorithms. c Comparison of SD values from positive genes (n = 52), background genes (n = 26, 044) and negative genes (n = 62) to positive genes. The y-axis indicates the SD value. Violin plots: median ± upper and lower quartiles; P values were calculated from two-sided Student’s t tests. d AUC values obtained from five different ML algorithms. For five algorithms, n = 20 independent replicates, respectively. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to span a 1.5 interquartile range from the edges. e Weighted SD value (SDw) of each gene in the candidate interval for 12 TBN QTLs. f Weighted probability (Pw) of each gene in 12 TBN QTLs, gray dotted line indicates the cutoff of machine learning. In e, f, Salmon solid dots indicate potential candidate genes, and gray solid dots indicate excluded genes.
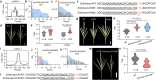
a Primary genetic mapping results of qTBN3-1. b Detailed weighted SD values (SDw) of genes in the interval of qTBN3-1. c Detailed weighted_Probability (Pw) of genes in the interval of qTBN3-1. d Representative photographs showing the TBN phenotype in wild type (left) and mutant Zmkinesin-EMS (right) derived from EMS materials. e TBN in wild type (gray violin plots) and mutants (pink violin plots). 20DHN, Hainan in winter 2020. 21CSD, Shandong in spring 2021. f Sequences of ZmKinesin target regions in wild type, Zmkinesin-KO#1 and Zmkinesin-KO#2 CRISPR/Cas9 knockout mutants. g Representative photographs showing the TBN phenotype in wild type (left), Zmkinesin-KO#1 (middle) and Zmkinesin-KO#2 (right) mutants. h TBN in wild type (left), Zmkinesin-KO#1 (middle) and Zmkinesin-KO#2 (right) mutants. i Primary genetic mapping results of qTBN7-1. j Detailed weighted SD values (SDw) of genes in the interval of qTBN7-1. k Detailed weighted_Probability (Pw) of genes in the interval of qTBN7-1. l Sequences of ZmHD-ZIP120 target regions in wild type, Zmhd-zip120-KO#1 CRISPR/Cas9 knockout mutants. m Representative photographs showing the TBN phenotype in wild type (left) and Zmhd-zip120-KO#1 (right) mutant. n TBN in wild type (left) and Zmhd-zip120-KO#1 (right) mutant. In b and j, Pink indicates SDd, light blue indicates SDv, and gray indicates SDg. In c and k, Pink indicates Pd, light blue indicates Pv, and gray indicates Pg. In f, l, the target sites and protospacer-adjacent motifs (PAM) are shown as underscored letters and pink letters, respectively. The gap lengths of sequences are shown above the wild type sequences. Scale bars referred above; 2 cm. In e, h, n, P values were determined by two-sided Student’s t tests. *P < 0.05, **P < 0.01, ***P < 0.001. Violin plots: median ± upper and lower quartiles. Source data are provided as a Source Data file.
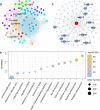
a Constructed network of TBN in maize. The seven genes identified in this study are indicated. Different colors indicated different biological pathways as follows: Hotpink, abscisic acid and reactive oxygen species; Royalblue, auxin; Lime, boundary; Brown, brassinosteroid; Cyan, cytokinin; Turquoise, cytoskeleton and cellulose; Magenta, flowering; Tan, gibberellin; Goldenrod, histone modification; Deepskyblue, meristem maintenance and determinacy; Orange, protein modification and transport; Blueviolet, sugar and nutrition. b Significant enrichment of 4278 network genes in multiple biological pathways. c Subnetwork of qTBN8-2 (lrs1).
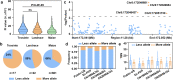
a Nucleotide diversity (pi value) across teosinte, landraces, and maize. n indicate 380, 387, 356 independent data points from left to right, respectively. ns, not significant. P values were determined by two-sided Student’s t tests. ***P < 0.001. Violin plots: median ± upper and lower quartiles. b Allele frequency of lrs1 across teosinte, landraces and maize. Less allele, allele associated with less TBN; More allele, allele associated with more TBN. c Association signals over lrs1 with TBN in a maize diversity panel. Four significant SNPs were uncovered. d Allele frequency of the SNP (Chr8: 172048437) in lrs1 across different modern breeding populations. e Variation in TBN across different populations, sorted as a function of the lead association SNP. n indicate 57, 13, 75, 8, 22, 6, 76, 15, 44, 7 independent inbred lines from left to right, respectively. Data represent means ± s.d.
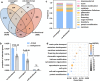
a Venn diagram showing the extent of overlap among node genes of the GRN, convergent selected genes only in MHGs, anti-directional selected genes, and co-directional selection genes between MHGs and FHGs. b Significant enrichments of convergent selected genes only in MHGs and co-directional selection genes between MHGs and FHGs in the constructed GRN. n indicate 100 independent experiments among three type background genes. Data represent means ± s.d. P values were determined by Chi-squared test. c Frequency of convergent selected genes only in MHGs and co-directional selection genes between MHGs and FHGs among 12 biological pathways. d Significant enrichment of 88 co-directional selection genes in multiple biological pathways.
Similar articles
-
Genetic Variation in ZmPAT7 Contributes to Tassel Branch Number in Maize.
Guan H, Chen X, Wang K, Liu X, Zhang D, Li Y, Song Y, Shi Y, Wang T, Li C, Li Y. Guan H, et al. Int J Mol Sci. 2022 Feb 26;23(5):2586. doi: 10.3390/ijms23052586. Int J Mol Sci. 2022. PMID: 35269730 Free PMC article.
-
Yang W, Zheng L, He Y, Zhu L, Chen X, Tao Y. Yang W, et al. Gene. 2020 Oct 5;757:144928. doi: 10.1016/j.gene.2020.144928. Epub 2020 Jul 2. Gene. 2020. PMID: 32622989
-
QDtbn1 , an F-box gene affecting maize tassel branch number by a dominant model.
Qin X, Tian S, Zhang W, Dong X, Ma C, Wang Y, Yan J, Yue B. Qin X, et al. Plant Biotechnol J. 2021 Jun;19(6):1183-1194. doi: 10.1111/pbi.13540. Epub 2021 Jan 19. Plant Biotechnol J. 2021. PMID: 33382512 Free PMC article.
-
Wang Y, Bao J, Wei X, Wu S, Fang C, Li Z, Qi Y, Gao Y, Dong Z, Wan X. Wang Y, et al. Cells. 2022 May 26;11(11):1753. doi: 10.3390/cells11111753. Cells. 2022. PMID: 35681448 Free PMC article. Review.
-
Li M, Zhong W, Yang F, Zhang Z. Li M, et al. Plant Cell Physiol. 2018 Mar 1;59(3):448-457. doi: 10.1093/pcp/pcy022. Plant Cell Physiol. 2018. PMID: 29420811 Review.
Cited by
-
Recent advances in exploring transcriptional regulatory landscape of crops.
Huo Q, Song R, Ma Z. Huo Q, et al. Front Plant Sci. 2024 Jun 5;15:1421503. doi: 10.3389/fpls.2024.1421503. eCollection 2024. Front Plant Sci. 2024. PMID: 38903438 Free PMC article. Review.
References
-
- Bazakos C, Hanemian M, Trontin C, Jimenez-Gomez JM, Loudet O. New strategies and tools in quantitative genetics: how to go from the phenotype to the genotype. Annu Rev. Plant Biol. 2017;68:435–455. - PubMed
-
- Jakobson CM, Jarosz DF. What has a century of quantitative genetics taught us about nature’s genetic tool kit? Annu. Rev. Genet. 2020;54:439–464. - PubMed
-
- Liang Y, Liu HJ, Yan J, Tian F. Natural variation in crops: realized understanding, continuing promise. Annu. Rev. Plant Biol. 2021;72:357–385. - PubMed
-
- Frary A, et al. fw2. 2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. - PubMed
-
- Thornsberry JM, et al. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001;28:286–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources