Signaling plasticity in the integrated stress response - PubMed
- ️Sun Jan 01 2023
Review
Signaling plasticity in the integrated stress response
Morgane Boone et al. Front Cell Dev Biol. 2023.
Abstract
The Integrated Stress Response (ISR) is an essential homeostatic signaling network that controls the cell's biosynthetic capacity. Four ISR sensor kinases detect multiple stressors and relay this information to downstream effectors by phosphorylating a common node: the alpha subunit of the eukaryotic initiation factor eIF2. As a result, general protein synthesis is repressed while select transcripts are preferentially translated, thus remodeling the proteome and transcriptome. Mounting evidence supports a view of the ISR as a dynamic signaling network with multiple modulators and feedback regulatory features that vary across cell and tissue types. Here, we discuss updated views on ISR sensor kinase mechanisms, how the subcellular localization of ISR components impacts signaling, and highlight ISR signaling differences across cells and tissues. Finally, we consider crosstalk between the ISR and other signaling pathways as a determinant of cell health.
Keywords: homeostasis; integrated stress response; sensor kinase; signal crosstalk; signal heterogeneity; signaling network; subcellular compartmentalization.
Copyright © 2023 Boone and Zappa.
Conflict of interest statement
Authors MB and FZ were employed by Altos labs.
Figures
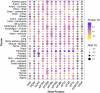
Tissue enrichment of core ISR components. Protein and RNA tissue specificity scores (TS) were extracted from the quantitative proteome map of the human body from the GTEx project, and are calculated as standard deviation from the bulk population average across all tissues for each gene product (Jiang et al., 2020). Grey dots: undetected by mass spectrometry. No data was available for ATF4 or HRI (EIF2AK1).
Similar articles
-
Surviving and Adapting to Stress: Translational Control and the Integrated Stress Response.
Wek RC, Anthony TG, Staschke KA. Wek RC, et al. Antioxid Redox Signal. 2023 Aug;39(4-6):351-373. doi: 10.1089/ars.2022.0123. Epub 2023 May 9. Antioxid Redox Signal. 2023. PMID: 36943285 Free PMC article. Review.
-
Mijit M, Boner M, Cordova RA, Gampala S, Kpenu E, Klunk AJ, Zhang C, Kelley MR, Staschke KA, Fishel ML. Mijit M, et al. Front Med (Lausanne). 2023 Apr 27;10:1146115. doi: 10.3389/fmed.2023.1146115. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37181357 Free PMC article.
-
Bond S, Lopez-Lloreda C, Gannon PJ, Akay-Espinoza C, Jordan-Sciutto KL. Bond S, et al. J Neuropathol Exp Neurol. 2020 Feb 1;79(2):123-143. doi: 10.1093/jnen/nlz129. J Neuropathol Exp Neurol. 2020. PMID: 31913484 Free PMC article. Review.
-
Mammalian integrated stress responses in stressed organelles and their functions.
Lu HJ, Koju N, Sheng R. Lu HJ, et al. Acta Pharmacol Sin. 2024 Jun;45(6):1095-1114. doi: 10.1038/s41401-023-01225-0. Epub 2024 Jan 24. Acta Pharmacol Sin. 2024. PMID: 38267546 Review.
-
The eIF2α kinases: their structures and functions.
Donnelly N, Gorman AM, Gupta S, Samali A. Donnelly N, et al. Cell Mol Life Sci. 2013 Oct;70(19):3493-511. doi: 10.1007/s00018-012-1252-6. Epub 2013 Jan 26. Cell Mol Life Sci. 2013. PMID: 23354059 Free PMC article. Review.
Cited by
-
Levin M. Levin M. Entropy (Basel). 2024 May 31;26(6):481. doi: 10.3390/e26060481. Entropy (Basel). 2024. PMID: 38920491 Free PMC article.
References
Publication types
Grants and funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received funding from Altos Labs Inc. The funder was not involved in the data collection, interpretation, and writing of this article or the decision to submit it for publication.
LinkOut - more resources
Full Text Sources
Research Materials